Elucidation of Pharmacological Mechanism Underlying the Anti-Alzheimer's Disease Effects of Evodia rutaecarpa and Discovery of Novel Lead Molecules: An In Silico Study
- PMID: 37570816
- PMCID: PMC10421504
- DOI: 10.3390/molecules28155846
Elucidation of Pharmacological Mechanism Underlying the Anti-Alzheimer's Disease Effects of Evodia rutaecarpa and Discovery of Novel Lead Molecules: An In Silico Study
Abstract
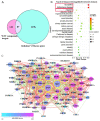
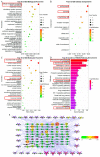
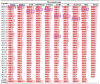
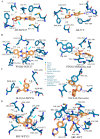
Alzheimer's disease (AD) is a brain disease with a peculiarity of multiformity and an insidious onset. Multiple-target drugs, especially Chinese traditional medicine, have achieved a measure of success in AD treatment. Evodia rutaecarpa (Juss.) Benth. (Wuzhuyu, WZY, i.e., E. rutaecarpa), a traditional Chinese herb, has been identified as an effective drug to cure migraines. To our surprise, our in silico study showed that rather than migraines, Alzheimer's disease was the primary disease to which the E. rutaecarpa active compounds were targeted. Correspondingly, a behavioral experiment showed that E. rutaecarpa extract could improve impairments in learning and memory in AD model mice. However, the mechanism underlying the way that E. rutaecarpa compounds target AD is still not clear. For this purpose, we employed methods of pharmacology networking and molecular docking to explore this mechanism. We found that E. rutaecarpa showed significant AD-targeting characteristics, and alkaloids of E. rutaecarpa played the main role in binding to the key nodes of AD. Our research detected that E. rutaecarpa affects the pathologic development of AD through the serotonergic synapse signaling pathway (SLC6A4), hormones (PTGS2, ESR1, AR), anti-neuroinflammation (SRC, TNF, NOS3), transcription regulation (NR3C1), and molecular chaperones (HSP90AA1), especially in the key nodes of PTGS2, AR, SLCA64, and SRC. Graveoline, 5-methoxy-N, N-dimethyltryptamine, dehydroevodiamine, and goshuyuamide II in E. rutaecarpa show stronger binding affinities to these key proteins than currently known preclinical and clinical drugs, showing a great potential to be developed as lead molecules for treating AD.
Keywords: Alzheimer’s disease; Evodia rutaecarpa; hormones; migraines; neurotransmitter inflammation; pharmacology network.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Anti-inflammatory effects and mechanisms of the ethanol extract of Evodia rutaecarpa and its bioactive components on neutrophils and microglial cells.Eur J Pharmacol. 2007 Jan 26;555(2-3):211-7. doi: 10.1016/j.ejphar.2006.10.002. Epub 2006 Oct 17. Eur J Pharmacol. 2007. PMID: 17109845
-
Bitterness and antibacterial activities of constituents from Evodia rutaecarpa.BMC Complement Altern Med. 2017 Mar 29;17(1):180. doi: 10.1186/s12906-017-1701-8. BMC Complement Altern Med. 2017. PMID: 28356098 Free PMC article.
-
Potential active compounds and common mechanisms of Evodia rutaecarpa for Alzheimer's disease comorbid pain by network pharmacology analysis.Heliyon. 2023 Jul 20;9(8):e18455. doi: 10.1016/j.heliyon.2023.e18455. eCollection 2023 Aug. Heliyon. 2023. PMID: 37529338 Free PMC article.
-
Therapeutic Effects of the major alkaloid constituents of Evodia rutaecarpa in Alzheimer's disease.Psychogeriatrics. 2024 Mar;24(2):443-457. doi: 10.1111/psyg.13051. Epub 2024 Jan 3. Psychogeriatrics. 2024. PMID: 38173117 Review.
-
Major Indole Alkaloids in Evodia Rutaecarpa: The Latest Insights and Review of Their Impact on Gastrointestinal Diseases.Biomed Pharmacother. 2023 Nov;167:115495. doi: 10.1016/j.biopha.2023.115495. Epub 2023 Sep 21. Biomed Pharmacother. 2023. PMID: 37741256 Review.
Cited by
-
A Natural Compound Containing a Disaccharide Structure of Glucose and Rhamnose Identified as Potential N-Glycanase 1 (NGLY1) Inhibitors.Molecules. 2023 Nov 24;28(23):7758. doi: 10.3390/molecules28237758. Molecules. 2023. PMID: 38067490 Free PMC article.
-
Selection of Reference Genes in Evodia rutaecarpa var. officinalis and Expression Patterns of Genes Involved in Its Limonin Biosynthesis.Plants (Basel). 2023 Sep 7;12(18):3197. doi: 10.3390/plants12183197. Plants (Basel). 2023. PMID: 37765365 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

