Identification of Bacterial Metabolites Modulating Breast Cancer Cell Proliferation and Epithelial-Mesenchymal Transition
- PMID: 37570868
- PMCID: PMC10420980
- DOI: 10.3390/molecules28155898
Identification of Bacterial Metabolites Modulating Breast Cancer Cell Proliferation and Epithelial-Mesenchymal Transition
Abstract
Breast cancer patients are characterized by the oncobiotic transformation of multiple microbiome communities, including the gut microbiome. Oncobiotic transformation of the gut microbiome impairs the production of antineoplastic bacterial metabolites. The goal of this study was to identify bacterial metabolites with antineoplastic properties. We constructed a 30-member bacterial metabolite library and screened the library compounds for effects on cell proliferation and epithelial-mesenchymal transition. The metabolites were applied to 4T1 murine breast cancer cells in concentrations corresponding to the reference serum concentrations. However, yric acid, glycolic acid, d-mannitol, 2,3-butanediol, and trans-ferulic acid exerted cytostatic effects, and 3-hydroxyphenylacetic acid, 4-hydroxybenzoic acid, and vanillic acid exerted hyperproliferative effects. Furthermore, 3-hydroxyphenylacetic acid, 4-hydroxybenzoic acid, 2,3-butanediol, and hydrocinnamic acid inhibited epithelial-to-mesenchymal (EMT) transition. We identified redox sets among the metabolites (d-mannitol-d-mannose, 1-butanol-butyric acid, ethylene glycol-glycolic acid-oxalic acid), wherein only one partner within the set (d-mannitol, butyric acid, glycolic acid) possessed bioactivity in our system, suggesting that changes to the local redox potential may affect the bacterial secretome. Of the nine bioactive metabolites, 2,3-butanediol was the only compound with both cytostatic and anti-EMT properties.
Keywords: 2,3-butanediol; 3-hydroxyphenylacetic acid; 4-hydroxybenzoic acid; bacterial metabolite; breast cancer; butyric acid; d-mannitol; dysbiosis; epithelial-mesenchymal transition; glycolic acid; high content screening; hydrocinnamic acid; metabolite signaling; microbiome; proliferation; secretome; trans-ferulic acid; vanillic acid.
Conflict of interest statement
Bai is a shareholder and CEO of Holobiont Diagnostics Ltd., which are involved in the development of microbiome-based diagnostic tools. Other authors declare no conflict of interest.
Figures


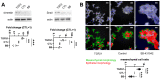
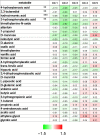


References
MeSH terms
Substances
Grants and funding
- K123975/National Research, Development and Innovation Office
- K124141/National Research, Development and Innovation Office
- FK128387/National Research, Development and Innovation Office
- TKP2021-EGA-19/National Research, Development and Innovation Office
- TKP2021-EGA-20/National Research, Development and Innovation Office
LinkOut - more resources
Full Text Sources
Medical

