Microfibrous Scaffolds Guide Stem Cell Lumenogenesis and Brain Organoid Engineering
- PMID: 37572376
- PMCID: PMC7617127
- DOI: 10.1002/adma.202300305
Microfibrous Scaffolds Guide Stem Cell Lumenogenesis and Brain Organoid Engineering
Abstract
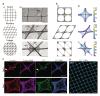
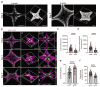
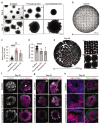
3D organoids are widely used as tractable in vitro models capable of elucidating aspects of human development and disease. However, the manual and low-throughput culture methods, coupled with a low reproducibility and geometric heterogeneity, restrict the scope and application of organoid research. Combining expertise from stem cell biology and bioengineering offers a promising approach to address some of these limitations. Here, melt electrospinning writing is used to generate tuneable grid scaffolds that can guide the self-organization of pluripotent stem cells into patterned arrays of embryoid bodies. Grid geometry is shown to be a key determinant of stem cell self-organization, guiding the position and size of emerging lumens via curvature-controlled tissue growth. Two distinct methods for culturing scaffold-grown embryoid bodies into either interconnected or spatially discrete cerebral organoids are reported. These scaffolds provide a high-throughput method to generate, culture, and analyze large numbers of organoids, substantially reducing the time investment and manual labor involved in conventional methods of organoid culture. It is anticipated that this methodological development will open up new opportunities for guiding pluripotent stem cell culture, studying lumenogenesis, and generating large numbers of uniform organoids for high-throughput screening.
Keywords: bioengineering; lumenogenesis; melt electrospinning writing; organoids; scaffolds; stem cells.
© 2023 The Authors. Advanced Materials published by Wiley-VCH GmbH.
Conflict of interest statement
K.I.R.R., R. W., R.X., J.P.K.A., C.L.G., D.R., J.A.K. and M.M.S. are coinventors on a patent describing methods for preparing organoids on lattice scaffolds. The rest of the authors declare no conflicts of interest.
Figures



References
-
- Bagnat M, Navis A. Cell Polarity 2: Role in Development and Disease. Springer International Publishing; New York: 2015. pp. 67–93.
-
- Sigurbjörnsdóttir S, Mathew R, Leptin M. Nat Rev Mol Cell Biol. 2014;15:665. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

