Loss of CD4+ T cell-intrinsic arginase 1 accelerates Th1 response kinetics and reduces lung pathology during influenza infection
- PMID: 37572656
- PMCID: PMC10576612
- DOI: 10.1016/j.immuni.2023.07.014
Loss of CD4+ T cell-intrinsic arginase 1 accelerates Th1 response kinetics and reduces lung pathology during influenza infection
Abstract
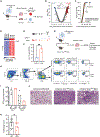
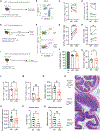
Arginase 1 (Arg1), the enzyme catalyzing the conversion of arginine to ornithine, is a hallmark of IL-10-producing immunoregulatory M2 macrophages. However, its expression in T cells is disputed. Here, we demonstrate that induction of Arg1 expression is a key feature of lung CD4+ T cells during mouse in vivo influenza infection. Conditional ablation of Arg1 in CD4+ T cells accelerated both virus-specific T helper 1 (Th1) effector responses and its resolution, resulting in efficient viral clearance and reduced lung pathology. Using unbiased transcriptomics and metabolomics, we found that Arg1-deficiency was distinct from Arg2-deficiency and caused altered glutamine metabolism. Rebalancing this perturbed glutamine flux normalized the cellular Th1 response. CD4+ T cells from rare ARG1-deficient patients or CRISPR-Cas9-mediated ARG1-deletion in healthy donor cells phenocopied the murine cellular phenotype. Collectively, CD4+ T cell-intrinsic Arg1 functions as an unexpected rheostat regulating the kinetics of the mammalian Th1 lifecycle with implications for Th1-associated tissue pathologies.
Keywords: IFN-γ; IL-10; Th1 immunity; arginase 1; arginase 1 deficiency; autoimmunity; cell metabolism; complement; glutamine; influenza infection.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests M.S. is an inventor on a patent describing the use of RBD ligands for cell-surface evaluation of CAT1/solute carrier family 7 member 1 (SLC7A1) and other solute carrier (SLC) expression (N.T. gave up her rights); he is a co-founder and head of the scientific board of METAFORA-Biosystems, a start-up company that focuses on metabolite transporters under physiological and pathological conditions.
Figures





References
-
- Gazzinelli RT, Wysocka M, Hieny S, Scharton-Kersten T, Cheever A, Kühn R, Muüller W, Trinchieri G, and Sher A. (1996). In the absence of endogenous IL-10, mice acutely infected with Toxoplasma gondii succumb to a lethal immune response dependent on CD4+ T cells and accompanied by overproduction of IL-12, IFN-gamma and TNF-alpha. J. Immunol 157, 798–805. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

