HDAC3 and HDAC8 PROTAC dual degrader reveals roles of histone acetylation in gene regulation
- PMID: 37572669
- PMCID: PMC10802846
- DOI: 10.1016/j.chembiol.2023.07.010
HDAC3 and HDAC8 PROTAC dual degrader reveals roles of histone acetylation in gene regulation
Abstract
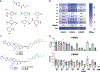
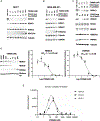
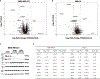
HDAC3 and HDAC8 have critical biological functions and represent highly sought-after therapeutic targets. Because histone deacetylases (HDACs) have a very conserved catalytic domain, developing isozyme-selective inhibitors remains challenging. HDAC3/8 also have deacetylase-independent activity, which cannot be blocked by conventional enzymatic inhibitors. Proteolysis-targeting chimeras (PROTACs) can selectively degrade a target enzyme, abolishing both enzymatic and scaffolding function. Here, we report a novel HDAC3/8 dual degrader YX968 that induces highly potent, rapid, and selective degradation of both HDAC3/8 without triggering pan-HDAC inhibitory effects. Unbiased quantitative proteomic experiments confirmed its high selectivity. HDAC3/8 degradation by YX968 does not induce histone hyperacetylation and broad transcriptomic perturbation. Thus, histone hyperacetylation may be a major factor for altering transcription. YX968 promotes apoptosis and kills cancer cells with a high potency in vitro. YX968 thus represents a new probe for dissecting the complex biological functions of HDAC3/8.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests X.Y., X.Z., G.Z., and D. Liao are co-inventors of a patent application related to this study filed on behalf of University of Florida Research Foundation. Other authors declare no competing interests.
Figures


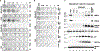
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

