Diversity and independent evolutionary profiling of rodent-borne viruses in Hainan, a tropical island of China
- PMID: 37572844
- PMCID: PMC10590688
- DOI: 10.1016/j.virs.2023.08.003
Diversity and independent evolutionary profiling of rodent-borne viruses in Hainan, a tropical island of China
Abstract
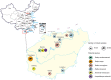
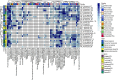
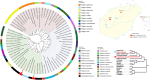
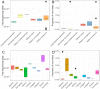
The risk of emerging infectious diseases (EID) is increasing globally. More than 60% of EIDs worldwide are caused by animal-borne pathogens. This study aimed to characterize the virome, analyze the phylogenetic evolution, and determine the diversity of rodent-borne viruses in Hainan Province, China. We collected 682 anal and throat samples from rodents, combined them into 28 pools according to their species and location, and processed them for next-generation sequencing and bioinformatics analysis. The diverse viral contigs closely related to mammals were assigned to 22 viral families. Molecular clues of the important rodent-borne viruses were further identified by polymerase chain reaction for phylogenetic analysis and annotation of genetic characteristics such as arenavirus, coronavirus, astrovirus, pestivirus, parvovirus, and papillomavirus. We identified pestivirus and bocavirus in Leopoldoms edwardsi from Huangjinjiaoling, and bocavirus in Rattus andamanensis from the national nature reserves of Bangxi with low amino acid identity to known pathogens are proposed as the novel species, and their rodent hosts have not been previously reported to carry these viruses. These results expand our knowledge of viral classification and host range and suggest that there are highly diverse, undiscovered viruses that have evolved independently in their unique wildlife hosts in inaccessible areas.
Keywords: Novel viruses; Phylogenetic analysis; Rodents; Virome; Wildlife.
Copyright © 2023 The Authors. Publishing services by Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest No potential conflict of interest was reported by the authors.
Figures





References
MeSH terms
LinkOut - more resources
Full Text Sources

