The copper-responsive regulator CsoR is indirectly involved in Bradyrhizobium diazoefficiens denitrification
- PMID: 37573143
- PMCID: PMC10457146
- DOI: 10.1093/femsle/fnad084
The copper-responsive regulator CsoR is indirectly involved in Bradyrhizobium diazoefficiens denitrification
Abstract
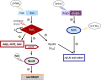
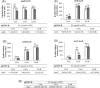
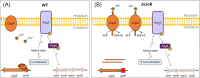
The soybean endosymbiont Bradyrhizobium diazoefficiens harbours the complete denitrification pathway that is catalysed by a periplasmic nitrate reductase (Nap), a copper (Cu)-containing nitrite reductase (NirK), a c-type nitric oxide reductase (cNor), and a nitrous oxide reductase (Nos), encoded by the napEDABC, nirK, norCBQD, and nosRZDFYLX genes, respectively. Induction of denitrification genes requires low oxygen and nitric oxide, both signals integrated into a complex regulatory network comprised by two interconnected cascades, FixLJ-FixK2-NnrR and RegSR-NifA. Copper is a cofactor of NirK and Nos, but it has also a role in denitrification gene expression and protein synthesis. In fact, Cu limitation triggers a substantial down-regulation of nirK, norCBQD, and nosRZDFYLX gene expression under denitrifying conditions. Bradyrhizobium diazoefficiens genome possesses a gene predicted to encode a Cu-responsive repressor of the CsoR family, which is located adjacent to copA, a gene encoding a putative Cu+-ATPase transporter. To investigate the role of CsoR in the control of denitrification gene expression in response to Cu, a csoR deletion mutant was constructed in this work. Mutation of csoR did not affect the capacity of B. diazoefficiens to grow under denitrifying conditions. However, by using qRT-PCR analyses, we showed that nirK and norCBQD expression was much lower in the csoR mutant compared to wild-type levels under Cu-limiting denitrifying conditions. On the contrary, copA expression was significantly increased in the csoR mutant. The results obtained suggest that CsoR acts as a repressor of copA. Under Cu limitation, CsoR has also an indirect role in the expression of nirK and norCBQD genes.
Keywords: Cu-responsive regulator; gene expression; nitric oxide reductase; nitrite reductase.
© The Author(s) 2023. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures


References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials

