High-density lipoprotein regulates angiogenesis by long non-coding RNA HDRACA
- PMID: 37574469
- PMCID: PMC10423722
- DOI: 10.1038/s41392-023-01558-6
High-density lipoprotein regulates angiogenesis by long non-coding RNA HDRACA
Abstract
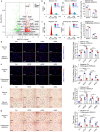
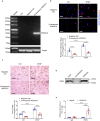
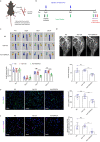
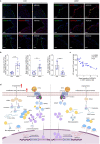
Normal high-density lipoprotein (nHDL) can induce angiogenesis in healthy individuals. However, HDL from patients with coronary artery disease undergoes various modifications, becomes dysfunctional (dHDL), and loses its ability to promote angiogenesis. Here, we identified a long non-coding RNA, HDRACA, that is involved in the regulation of angiogenesis by HDL. In this study, we showed that nHDL downregulates the expression of HDRACA in endothelial cells by activating WW domain-containing E3 ubiquitin protein ligase 2, which catalyzes the ubiquitination and subsequent degradation of its transcription factor, Kruppel-like factor 5, via sphingosine 1-phosphate (S1P) receptor 1. In contrast, dHDL with lower levels of S1P than nHDL were much less effective in decreasing the expression of HDRACA. HDRACA was able to bind to Ras-interacting protein 1 (RAIN) to hinder the interaction between RAIN and vigilin, which led to an increase in the binding between the vigilin protein and proliferating cell nuclear antigen (PCNA) mRNA, resulting in a decrease in the expression of PCNA and inhibition of angiogenesis. The expression of human HDRACA in a hindlimb ischemia mouse model inhibited the recovery of angiogenesis. Taken together, these findings suggest that HDRACA is involved in the HDL regulation of angiogenesis, which nHDL inhibits the expression of HDRACA to induce angiogenesis, and that dHDL is much less effective in inhibiting HDRACA expression, which provides an explanation for the decreased ability of dHDL to stimulate angiogenesis.
© 2023. West China Hospital, Sichuan University.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Toyota E, et al. Vascular endothelial growth factor is required for coronary collateral growth in the rat. Circulation. 2005;112:2108–2113. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

