The landscape of the immunoglobulin repertoire in endemic pemphigus foliaceus
- PMID: 37575223
- PMCID: PMC10421657
- DOI: 10.3389/fimmu.2023.1189251
The landscape of the immunoglobulin repertoire in endemic pemphigus foliaceus
Abstract
Introduction: Primarily driven by autoreactive B cells, pemphigus foliaceus (PF) is an uncommon autoimmune blistering skin disease of sporadic occurrence worldwide. However, PF reaches a prevalence of 3% in the endemic areas of Brazil, the highest ever registered for any autoimmune disease, which indicates environmental factors influencing the immune response in susceptible individuals. We aimed to provide insights into the immune repertoire of patients with PF living in the endemic region of the disease, compared to healthy individuals from the endemic region and a non-endemic area.
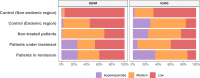
Methods: We characterized the B-cell repertoire in i) nontreated patients (n=5); ii) patients under immunosuppressive treatment (n=5); iii) patients in remission without treatment (n=6); and two control groups iv) from the endemic (n=6) and v) non-endemic areas in Brazil (n=4). We used total RNA extracted from peripheral blood mononuclear cells and performed a comprehensive characterization of the variable region of immunoglobulin heavy chain (IGH) in IgG and IgM using next-generation sequencing.
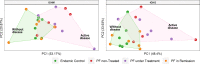
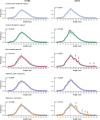
Results: Compared to individuals from a different area, we observed remarkably lower clonotype diversity in the B-cell immune repertoire of patients and controls from the endemic area (p < 0.02), suggesting that the immune repertoire in the endemic area is under geographically specific and intense environmental pressure. Moreover, we observed longer CDR3 sequences in patients, and we identified differential disease-specific usage of IGHV segments, including increased IGHV3-30 and decreased IGHV3-23 in patients with active disease (p < 0.04). Finally, our robust network analysis discovered clusters of CDR3 sequences uniquely observed in patients with PF.
Discussion: Our results indicate that environmental factors, in addition to disease state, impact the characteristics of the repertoire. Our findings can be applied to further investigation of the environmental factors that trigger pemphigus and expand the knowledge for identifying new targeted and more effective therapies.
Keywords: B cells; autoimmunity; environmental factors; immunoglobulin repertoire; pemphigus foliaceus; skin disease.
Copyright © 2023 Calonga-Solís, Olbrich, Ott, Adelman Cipolla, Malheiros, Künstner, Farias, Camargo, Petzl-Erler, Busch, Fähnrich and Augusto.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

