Characterization of the TRPV6 calcium channel-specific phenotype by RNA-seq in castration-resistant human prostate cancer cells
- PMID: 37576552
- PMCID: PMC10415680
- DOI: 10.3389/fgene.2023.1215645
Characterization of the TRPV6 calcium channel-specific phenotype by RNA-seq in castration-resistant human prostate cancer cells
Abstract
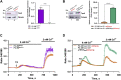
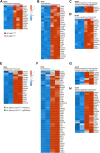
Background: Transient receptor potential vanilloid subfamily member 6 (TRPV6), a highly calcium-selective channel, has been shown to play a significant role in calcium homeostasis and to participate both in vitro and in vivo in growth, cell survival, and drug resistance of prostate cancer. Its role and the corresponding calcium-dependent pathways were mainly studied in hormone-dependent human prostate cancer cell lines, often used as a model of early-stage prostate cancers. The goal of the present study was to describe the TRPV6-specific phenotype and signaling pathways it is involved in, using castration-resistant prostate cancer cell lines. Methods: RNA sequencing (RNA-seq) was used to study the gene expression impacted by TRPV6 using PC3Mtrpv6-/- versus PC3Mtrpv6+/+ and its derivative PC3M-luc-C6trpv6+/+ cell line in its native and TRPV6 overexpressed form. In addition to the whole-cell RNA sequencing, immunoblotting, quantitative PCR, and calcium imaging were used to validate trpv6 gene status and functional consequences, in both trpv6 -/- and TRPV6 overexpression cell lines. Results: trpv6 -/- status was validated using both immunoblotting and quantitative PCR, and the functional consequences of either trpv6 gene deletion or TRPV6 overexpression were shown using calcium imaging. RNA-seq analysis demonstrated that the calcium channel TRPV6, being a crucial player of calcium signaling, significantly impacts the expression of genes involved in cancer progression, such as cell cycle regulation, chemotaxis, migration, invasion, apoptosis, ferroptosis as well as drug resistance, and extracellular matrix (ECM) re-organization. Conclusion: Our data suggest that the trpv6 gene is involved in and regulates multiple pathways related to tumor progression and drug resistance in castration-resistant prostate cancer cells.
Keywords: RNA-seq analysis; TRPV6 channel; castration-resistant prostate cancer; expression profile; signaling pathway.
Copyright © 2023 Cordier, Haustrate, Prevarskaya and Lehen’kyi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

