A simple sequence repeats marker of disease resistance in shrimp Litopenaeus vannamei and its application in selective breeding
- PMID: 37576558
- PMCID: PMC10415038
- DOI: 10.3389/fgene.2023.1144361
A simple sequence repeats marker of disease resistance in shrimp Litopenaeus vannamei and its application in selective breeding
Abstract
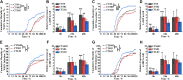
The polymorphism of the simple sequence repeat (SSR) in the 5' untranslated coding region (5'-UTR) of the antiviral gene IRF (LvIRF) has been shown to be implicated in the resistance to viral pathogens in shrimp Litopenaeus vannamei (L. vannamei). In this study, we explored the potential of this (CT)n-SSR marker in disease resistance breeding and the hereditary property of disease resistance traits in offspring. From 2018 to 2021, eight populations were generated through crossbreeding by selecting individuals according to microsatellite genotyping. Our results demonstrated that shrimp with the shorter (CT)n repeat exhibited higher resistance to white spot syndrome virus (WSSV) or Decapod iridescent virus 1 (DIV1); meanwhile, these resistance traits could be inherited in offspring. Interestingly, we observed that the longer (CT)n repeats were associated with bacterial resistance traits. Accordingly, shrimp with longer (CT)n repeats exhibited higher tolerance to Vibrio parahaemolyticus infection. Taken together, these results indicate that the single (CT)n-SSR marker could be used to selective breeding for both resistance to virus and bacteria in shrimps.
Keywords: IRF; disease resistance; selective breeding; shrimp; simple sequence repeat (SSR).
Copyright © 2023 Yin, Wang, Weng, Li, He and Li.
Conflict of interest statement
Author SL was employed by Guangdong Evergreen Feed Industry Co. Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Chakrabarty U., Dutta S., Mallik A., Mondal D., Mandal N. (2015). Identification and characterisation of microsatellite DNA markers in order to recognise the WSSV susceptible populations of marine giant black tiger shrimp, Penaeus monodon . Vet. Res. 46, 110. PMID 26407974. 10.1186/s13567-015-0248-2 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

