Evolutionary potential of the monkeypox genome arising from interactions with human APOBEC3 enzymes
- PMID: 37577211
- PMCID: PMC10422979
- DOI: 10.1093/ve/vead047
Evolutionary potential of the monkeypox genome arising from interactions with human APOBEC3 enzymes
Abstract
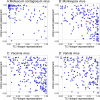
APOBEC3, an enzyme subfamily that plays a role in virus restriction by generating mutations at particular DNA motifs or mutational 'hotspots', can drive viral mutagenesis with host-specific preferential hotspot mutations contributing to pathogen variation. While previous analysis of viral genomes from the 2022 Mpox (formerly Monkeypox) disease outbreak has shown a high frequency of C>T mutations at TC motifs, suggesting recent mutations are human APOBEC3-mediated, how emerging monkeypox virus (MPXV) strains will evolve as a consequence of APOBEC3-mediated mutations remains unknown. By measuring hotspot under-representation, depletion at synonymous sites, and a combination of the two, we analyzed APOBEC3-driven evolution in human poxvirus genomes, finding varying hotspot under-representation patterns. While the native poxvirus molluscum contagiosum exhibits a signature consistent with extensive coevolution with human APOBEC3, including depletion of TC hotspots, variola virus shows an intermediate effect consistent with ongoing evolution at the time of eradication. MPXV, likely the result of recent zoonosis, showed many genes with more TC hotspots than expected by chance (over-representation) and fewer GC hotspots than expected (under-representation). These results suggest the MPXV genome: (1) may have evolved in a host with a particular APOBEC GC hotspot preference, (2) has inverted terminal repeat (ITR) regions-which may be exposed to APOBEC3 for longer during viral replication-and longer genes likely to evolve faster, and therefore (3) has a heightened potential for future human APOBEC3-meditated evolution as the virus spreads in the human population. Our predictions of MPXV mutational potential can both help guide future vaccine development and identification of putative drug targets and add urgency to the task of containing human Mpox disease transmission and uncovering the ecology of the virus in its reservoir host.
Keywords: APOBEC; computational biology; evolution; mokeypox; virus.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures



Update of
-
Evolutionary potential of the monkeypox genome arising from interactions with human APOBEC3 enzymes.bioRxiv [Preprint]. 2023 Jun 28:2023.06.27.546779. doi: 10.1101/2023.06.27.546779. bioRxiv. 2023. Update in: Virus Evol. 2023 Aug 02;9(2):vead047. doi: 10.1093/ve/vead047. PMID: 37425914 Free PMC article. Updated. Preprint.
References
-
- Alcamí A. et al. (1998) ‘Blockade of Chemokine Activity by a Soluble Chemokine Binding Protein from Vaccinia Virus’, The Journal of Immunology, 160: 624–33. - PubMed
-
- Brinkmann A. et al. (2022) ‘Possible Adaption of the 2022 Monkeypox Virus to the Human Host through Gene Duplication and Loss’, bioRxiv, 2022.10.21.512875.
-
- Burki T. (2023) ‘The End of the Mpox Pandemic?’, The Lancet Infectious Diseases, 23: 159–60. - PubMed
-
- Carvalho T. (2022) ‘The Unknown Efficacy of Tecovirimat against Monkeypox’, Nature Medicine, 28: 2224–5. - PubMed

