Improving taxonomic classification with feature space balancing
- PMID: 37577265
- PMCID: PMC10415173
- DOI: 10.1093/bioadv/vbad092
Improving taxonomic classification with feature space balancing
Abstract
Summary: Modern high-throughput sequencing technologies, such as metagenomic sequencing, generate millions of sequences that need to be assigned to their taxonomic rank. Modern approaches either apply local alignment to existing databases, such as MMseqs2, or use deep neural networks, as in DeepMicrobes and BERTax. Due to the increasing size of datasets and databases, alignment-based approaches are expensive in terms of runtime. Deep learning-based approaches can require specialized hardware and consume large amounts of energy. In this article, we propose to use k-mer profiles of DNA sequences as features for taxonomic classification. Although k-mer profiles have been used before, we were able to significantly increase their predictive power significantly by applying a feature space balancing approach to the training data. This greatly improved the generalization quality of the classifiers. We have implemented different pipelines using our proposed feature extraction and dataset balancing in combination with different simple classifiers, such as bagged decision trees or feature subspace KNNs. By comparing the performance of our pipelines with state-of-the-art algorithms, such as BERTax and MMseqs2 on two different datasets, we show that our pipelines outperform these in almost all classification tasks. In particular, sequences from organisms that were not part of the training were classified with high precision.
Availability and implementation: The open-source code and the code to reproduce the results is available in Seafile, at https://tinyurl.com/ysk47fmr.
Supplementary information: Supplementary data are available at Bioinformatics Advances online.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
The authors have no conflicts of interest to declare. All authors have seen and agree with the contents of the manuscript and there is no financial interest to report.
Figures
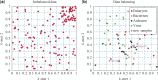
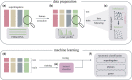


References
LinkOut - more resources
Full Text Sources
Miscellaneous

