Evolution of primate interferon-induced transmembrane proteins (IFITMs): a story of gain and loss with a differentiation into a canonical cluster and IFITM retrogenes
- PMID: 37577422
- PMCID: PMC10415907
- DOI: 10.3389/fmicb.2023.1213685
Evolution of primate interferon-induced transmembrane proteins (IFITMs): a story of gain and loss with a differentiation into a canonical cluster and IFITM retrogenes
Abstract
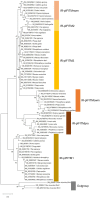
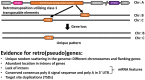
Interferon-inducible transmembrane proteins (IFITMs) are a family of transmembrane proteins. The subgroup of immunity-related (IR-)IFITMs is involved in adaptive and innate immune responses, being especially active against viruses. Here, we suggest that IFITMs should be classified as (1) a canonical IFITM gene cluster, which is located on the same chromosome, and (2) IFITM retrogenes, with a random and unique location at different positions within the genome. Phylogenetic analyses of the canonical cluster revealed the existence of three novel groups of primate IFITMs (pIFITM) in the IR-IFITM clade: the prosimian pIFITMs(pro), the new world monkey pIFITMs(nwm) and the old world monkey pIFITMs(owm). Therefore, we propose a new nomenclature: IR-pIFITM1, IR-pIFITM2, IR-pIFITM3, IR-pIFITMnwm, IR-pIFITMowm, and IR-pIFITMpro. We observed divergent evolution for pIFITM5 and pIFITM10, and evidence for concerted evolution and a mechanism of birth-and-death evolution model for the IR-pIFITMs. In contrast, the IFITMs scattered throughout the genomes possessed features of retrogenes retrotransposed by class 1 transposable elements. The origin of the IFITM retrogenes correspond to more recent events. We hypothesize that the transcript of a canonical IFITM3 has been constantly retrotransposed using class 1 transposable elements resulting in the IFITM retro(pseudo)genes. The unique pattern of each species has most likely been caused by constant pseudogenization and loss of the retro(pseudo)genes. This suggests a third mechanism of evolution for the IR-IFITMs in primates, similar to the birth-and-death model of evolution, but via a transposable element mechanism, which resulted in retro(pseudo)genes.
Keywords: antiviral proteins; evolution; innate immunity; interferon-induced transmembrane proteins; primates; retrogene; transposable elements.
Copyright © 2023 Schelle, Abrantes, Baldauf and Esteves.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Functional Heterogeneity of Mammalian IFITM Proteins against HIV-1.J Virol. 2021 Aug 25;95(18):e0043921. doi: 10.1128/JVI.00439-21. Epub 2021 Aug 25. J Virol. 2021. PMID: 34160255 Free PMC article.
-
Nonhuman Primate IFITM Proteins Are Potent Inhibitors of HIV and SIV.PLoS One. 2016 Jun 3;11(6):e0156739. doi: 10.1371/journal.pone.0156739. eCollection 2016. PLoS One. 2016. PMID: 27257969 Free PMC article.
-
Ferret Interferon (IFN)-Inducible Transmembrane Proteins Are Upregulated by both IFN-α and Influenza Virus Infection.J Virol. 2021 Jun 24;95(14):e0011121. doi: 10.1128/JVI.00111-21. Epub 2021 Jun 24. J Virol. 2021. PMID: 33952646 Free PMC article.
-
Functional Involvement of Interferon-Inducible Transmembrane Proteins in Antiviral Immunity.Front Microbiol. 2019 May 16;10:1097. doi: 10.3389/fmicb.2019.01097. eCollection 2019. Front Microbiol. 2019. PMID: 31156602 Free PMC article. Review.
-
IFITM proteins: Understanding their diverse roles in viral infection, cancer, and immunity.J Biol Chem. 2023 Jan;299(1):102741. doi: 10.1016/j.jbc.2022.102741. Epub 2022 Nov 23. J Biol Chem. 2023. PMID: 36435199 Free PMC article. Review.
Cited by
-
Alternative splicing expands the antiviral IFITM repertoire in Chinese rufous horseshoe bats.PLoS Pathog. 2024 Dec 26;20(12):e1012763. doi: 10.1371/journal.ppat.1012763. eCollection 2024 Dec. PLoS Pathog. 2024. PMID: 39724110 Free PMC article.
-
Role of IFITM2 in osteogenic differentiation of C3H10T1/2 mesenchymal stem cells.Intractable Rare Dis Res. 2024 Feb;13(1):42-50. doi: 10.5582/irdr.2023.01108. Intractable Rare Dis Res. 2024. PMID: 38404731 Free PMC article.
-
Revealing Fibrosis Genes as Biomarkers of Ulcerative Colitis: A Bioinformatics Study Based on ScRNA and Bulk RNA Datasets.Endocr Metab Immune Disord Drug Targets. 2025;25(9):710-720. doi: 10.2174/0118715303332155240912050838. Endocr Metab Immune Disord Drug Targets. 2025. PMID: 39428941
References
LinkOut - more resources
Full Text Sources

