This is a preprint.
CCT and Cullin1 regulate the TORC1 pathway to promote dendritic arborization in health and disease
- PMID: 37577581
- PMCID: PMC10418059
- DOI: 10.1101/2023.07.31.551324
CCT and Cullin1 regulate the TORC1 pathway to promote dendritic arborization in health and disease
Update in
-
CCT and Cullin1 Regulate the TORC1 Pathway to Promote Dendritic Arborization in Health and Disease.Cells. 2024 Jun 13;13(12):1029. doi: 10.3390/cells13121029. Cells. 2024. PMID: 38920658 Free PMC article.
Abstract
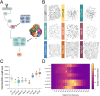
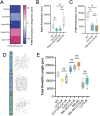
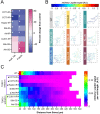
The development of cell-type-specific dendritic arbors is integral to the proper functioning of neurons within their circuit networks. In this study, we examine the regulatory relationship between the cytosolic chaperonin CCT, key insulin pathway genes, and an E3 ubiquitin ligase (Cullin1) in homeostatic dendritic development. CCT loss of function (LOF) results in dendritic hypotrophy in Drosophila Class IV (CIV) multidendritic larval sensory neurons, and CCT has recently been shown to fold components of the TOR (Target of Rapamycin) complex 1 (TORC1), in vitro. Through targeted genetic manipulations, we have confirmed that LOF of CCT and the TORC1 pathway reduces dendritic complexity, while overexpression of key TORC1 pathway genes increases dendritic complexity in CIV neurons. Both CCT and TORC1 LOF significantly reduce microtubule (MT) stability. CCT has been previously implicated in regulating proteinopathic aggregation, thus we examined CIV dendritic development in disease conditions as well. Expression of mutant Huntingtin leads to dendritic hypotrophy in a repeat-length-dependent manner, which can be rescued by TORC1 disinhibition via Cullin1 LOF. Together, our data suggest that Cullin1 and CCT influence dendritic arborization through regulation of TORC1 in both health and disease.
Conflict of interest statement
CONFLICT OF INTEREST The authors declare no competing financial interests.
Figures

References
-
- Arshadi C, Günther U, Eddison M, Harrington KIS, Ferreira TA (2021) SNT: a unifying toolbox for quantification of neuronal anatomy. Nat Methods 18:374–377. - PubMed
-
- Barnat M, Le Friec J, Benstaali C, Humbert S (2017) Huntingtin-Mediated Multipolar-Bipolar Transition of Newborn Cortical Neurons Is Critical for Their Postnatal Neuronal Morphology. Neuron 93:99–114. - PubMed
-
- Berger Z, Ravikumar B, Menzies FM, Garcia Oroz L, Underwood BR, Pangalos MN, Schmitt I, Wullner U, Evert BO, O’kane CJ, Rubinsztein DC (2006) Rapamycin alleviates toxicity of different aggregate-prone proteins. Human Molecular Genetics 15:433–442. - PubMed
-
- Bertrand M, Decoville M, Meudal H, Birman S, Landon C (2020) Metabolomic nuclear magnetic resonance studies at presymptomatic and symptomatic stages of huntington’s disease on a drosophila model. Journal of Proteome Research 19:4034–4045. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
