This is a preprint.
Requirement of GrgA for Chlamydia infectious progeny production, optimal growth, and efficient plasmid maintenance
- PMID: 37577610
- PMCID: PMC10418237
- DOI: 10.1101/2023.08.02.551707
Requirement of GrgA for Chlamydia infectious progeny production, optimal growth, and efficient plasmid maintenance
Update in
-
Requirement of GrgA for Chlamydia infectious progeny production, optimal growth, and efficient plasmid maintenance.mBio. 2024 Jan 16;15(1):e0203623. doi: 10.1128/mbio.02036-23. Epub 2023 Dec 19. mBio. 2024. PMID: 38112466 Free PMC article.
Abstract
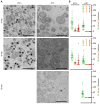
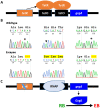
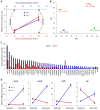
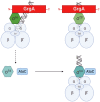
Chlamydia, an obligate intracellular bacterial pathogen, has a unique developmental cycle involving the differentiation of invading elementary bodies (EBs) to noninfectious reticulate bodies (RBs), replication of RBs, and redifferentiation of RBs into progeny EBs. Progression of this cycle is regulated by three sigma factors, which direct the RNA polymerase to their respective target gene promoters. We hypothesized that the Chlamydia-specific transcriptional regulator GrgA, previously shown to activate σ66 and σ28, plays an essential role in chlamydial development and growth. To test this hypothesis, we applied a novel genetic tool known as dependence on plasmid-mediated expression (DOPE) to create Chlamydia trachomatis with conditional GrgA-deficiency. We show that GrgA-deficient C. trachomatis RBs have a growth rate that is approximately half of the normal rate and fail to transition into progeny EBs. In addition, GrgA-deficient C. trachomatis fail to maintain its virulence plasmid. Results of RNA-seq analysis indicate that GrgA promotes RB growth by optimizing tRNA synthesis and expression of nutrient-acquisition genes, while it enables RB-to-EB conversion by facilitating the expression of a histone and outer membrane proteins required for EB morphogenesis. GrgA also regulates numerous other late genes required for host cell exit and subsequent EB invasion into host cells. Importantly, GrgA stimulates the expression of σ54, the third and last sigma factor, and its activator AtoC, and thereby indirectly upregulating the expression of σ54-dependent genes. In conclusion, our work demonstrates that GrgA is a master transcriptional regulator in Chlamydia and plays multiple essential roles in chlamydial pathogenicity.
Conflict of interest statement
COMPETING INTERESTS STATEMENT The authors declare no conflict of interest.
Figures




Similar articles
-
Requirement of GrgA for Chlamydia infectious progeny production, optimal growth, and efficient plasmid maintenance.mBio. 2024 Jan 16;15(1):e0203623. doi: 10.1128/mbio.02036-23. Epub 2023 Dec 19. mBio. 2024. PMID: 38112466 Free PMC article.
-
GrgA overexpression inhibits Chlamydia trachomatis growth through sigma66- and sigma28-dependent mechanisms.Microb Pathog. 2021 Jul;156:104917. doi: 10.1016/j.micpath.2021.104917. Epub 2021 May 1. Microb Pathog. 2021. PMID: 33940135 Free PMC article.
-
Identification of a GrgA-Euo-HrcA Transcriptional Regulatory Network in Chlamydia.mSystems. 2021 Aug 31;6(4):e0073821. doi: 10.1128/mSystems.00738-21. Epub 2021 Aug 3. mSystems. 2021. PMID: 34342542 Free PMC article.
-
Type III Secretion in Chlamydia.Microbiol Mol Biol Rev. 2023 Sep 26;87(3):e0003423. doi: 10.1128/mmbr.00034-23. Epub 2023 Jun 26. Microbiol Mol Biol Rev. 2023. PMID: 37358451 Free PMC article. Review.
-
The intracellular life of chlamydiae.Semin Pediatr Infect Dis. 2002 Oct;13(4):239-48. doi: 10.1053/spid.2002.127201. Semin Pediatr Infect Dis. 2002. PMID: 12491229 Review.
References
-
- Abdelrahman YM, Belland RJ. 2005. The chlamydial developmental cycle. FEMS Microbiology Reviews 29:949–59. - PubMed
-
- Lambden PR, Everson JS, Ward ME, Clarke IN. 1990. Sulfur-rich proteins of Chlamydia trachomatis: developmentally regulated transcription of polycistronic mRNA from tandem promoters. Gene 87:105–12. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
