This is a preprint.
ATAC and SAGA coactivator complexes utilize co-translational assembly, but their cellular localization properties and functions are distinct
- PMID: 37577620
- PMCID: PMC10418265
- DOI: 10.1101/2023.08.03.551787
ATAC and SAGA coactivator complexes utilize co-translational assembly, but their cellular localization properties and functions are distinct
Update in
-
ATAC and SAGA co-activator complexes utilize co-translational assembly, but their cellular localization properties and functions are distinct.Cell Rep. 2023 Sep 26;42(9):113099. doi: 10.1016/j.celrep.2023.113099. Epub 2023 Sep 8. Cell Rep. 2023. PMID: 37682711 Free PMC article.
Abstract
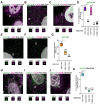
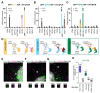
To understand the function of multisubunit complexes it is of key importance to uncover the precise mechanisms that guide their assembly. Nascent proteins can find and bind their interaction partners during their translation, leading to co-translational assembly. Here we demonstrate that the core modules of ATAC (ADA-Two-A-Containing) and SAGA (Spt-Ada-Gcn5-acetyltransferase), two lysine acetyl transferase-containing transcription coactivator complexes, assemble co-translationally in the cytoplasm of mammalian cells. In addition, SAGA complex containing all of its modules forms in the cytoplasm and acetylates non-histones proteins. In contrast, fully assembled ATAC complex cannot be detected in the cytoplasm of mammalian cells. However, endogenous ATAC complex containing two functional modules forms and functions in the nucleus. Thus, the two related coactivators, ATAC and SAGA, assemble by using co-translational pathways, but their subcellular localization, cytoplasmic abundance and functions are distinct.
Conflict of interest statement
Declaration of Interest
The authors do not declare conflict of interest.
Figures




References
-
- Riss A., Scheer E., Joint M., Trowitzsch S., Berger I., and Tora L. (2015). Subunits of ADA-two-A-containing (ATAC) or Spt-Ada-Gcn5-acetyltrasferase (SAGA) Coactivator Complexes Enhance the Acetyltransferase Activity of GCN5. The Journal of biological chemistry 290, 28997–29009. 10.1074/jbc.M115.668533. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
