In-silico and in-vitro morphometric analysis of intestinal organoids
- PMID: 37578984
- PMCID: PMC10473498
- DOI: 10.1371/journal.pcbi.1011386
In-silico and in-vitro morphometric analysis of intestinal organoids
Abstract
Organoids offer a powerful model to study cellular self-organisation, the growth of specific tissue morphologies in-vitro, and to assess potential medical therapies. However, the intrinsic mechanisms of these systems are not entirely understood yet, which can result in variability of organoids due to differences in culture conditions and basement membrane extracts used. Improving the standardisation of organoid cultures is essential for their implementation in clinical protocols. Developing tools to assess and predict the behaviour of these systems may produce a more robust and standardised biological model to perform accurate clinical studies. Here, we developed an algorithm to automate crypt-like structure counting on intestinal organoids in both in-vitro and in-silico images. In addition, we modified an existing two-dimensional agent-based mathematical model of intestinal organoids to better describe the system physiology, and evaluated its ability to replicate budding structures compared to new experimental data we generated. The crypt-counting algorithm proved useful in approximating the average number of budding structures found in our in-vitro intestinal organoid culture images on days 3 and 7 after seeding. Our changes to the in-silico model maintain the potential to produce simulations that replicate the number of budding structures found on days 5 and 7 of in-vitro data. The present study aims to aid in quantifying key morphological structures and provide a method to compare both in-vitro and in-silico experiments. Our results could be extended later to 3D in-silico models.
Copyright: © 2023 Montes-Olivas et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

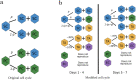
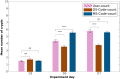
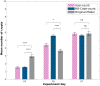
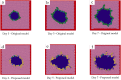
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

