Evolutionary history of MEK1 illuminates the nature of deleterious mutations
- PMID: 37579140
- PMCID: PMC10450672
- DOI: 10.1073/pnas.2304184120
Evolutionary history of MEK1 illuminates the nature of deleterious mutations
Abstract
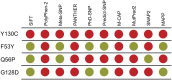
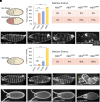
Mutations in signal transduction pathways lead to various diseases including cancers. MEK1 kinase, encoded by the human MAP2K1 gene, is one of the central components of the MAPK pathway and more than a hundred somatic mutations in the MAP2K1 gene were identified in various tumors. Germline mutations deregulating MEK1 also lead to congenital abnormalities, such as the cardiofaciocutaneous syndrome and arteriovenous malformation. Evaluating variants associated with a disease is a challenge, and computational genomic approaches aid in this process. Establishing evolutionary history of a gene improves computational prediction of disease-causing mutations; however, the evolutionary history of MEK1 is not well understood. Here, by revealing a precise evolutionary history of MEK1, we construct a well-defined dataset of MEK1 metazoan orthologs, which provides sufficient depth to distinguish between conserved and variable amino acid positions. We matched known and predicted disease-causing and benign mutations to evolutionary changes observed in corresponding amino acid positions and found that all known and many suspected disease-causing mutations are evolutionarily intolerable. We selected several variants that cannot be unambiguously assessed by automated prediction tools but that are confidently identified as "damaging" by our approach, for experimental validation in Drosophila. In all cases, evolutionary intolerant variants caused increased mortality and severe defects in fruit fly embryos confirming their damaging nature. We anticipate that our analysis will serve as a blueprint to help evaluate known and novel missense variants in MEK1 and that our approach will contribute to improving automated tools for disease-associated variant interpretation.
Keywords: Drosophila; damaging mutations; serine–threonine kinase; variants of unknown significance.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

