The isolation of rumen enterococci strains along with high potential utilizing cyanide
- PMID: 37580363
- PMCID: PMC10425440
- DOI: 10.1038/s41598-023-40488-9
The isolation of rumen enterococci strains along with high potential utilizing cyanide
Abstract
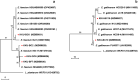
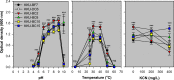
Cyanogenic glycosides in forage species and the possibility of cyanide (CN) poisoning can have undesirable effects on ruminants. The literature estimates that unknown rumen bacteria with rhodanese activity are key factors in the animal detoxification of cyanogenic glycosides, as they are capable of transforming CN into the less toxic thiocyanate. Therefore, identifying these bacteria will enhance our understanding of how to improve animal health with this natural CN detoxification process. In this study, a rhodanese activity screening assay revealed 6 of 44 candidate rumen bacterial strains isolated from domestic buffalo, dairy cattle, and beef cattle, each with a different colony morphology. These strains were identified as belonging to the species Enterococcus faecium and E. gallinarum by 16S ribosomal DNA sequence analysis. A CN-thiocyanate transformation assay showed that the thiocyanate formation capacity of the strains after a 12 h incubation ranged from 4.42 to 25.49 mg hydrogen CN equivalent/L. In addition, thiocyanate degradation resulted in the production of ammonia nitrogen and acetic acid in different strains. This study showed that certain strains of enterococci substantially contribute to CN metabolism in ruminants. Our results may serve as a starting point for research aimed at improving ruminant production systems in relation to CN metabolism.
© 2023. Springer Nature Limited.
Conflict of interest statement
S.K. owns Animal Feed Inter Trade Co. Ltd. that may potentially benefit from the research findings. All authors declare no completing interests.
Figures



References
-
- McSweeney CS, Odenyo A, Krause DO. Rumen microbial responses to antinutritive factors in fodder trees and shrub legumes. J. Appl. Anim. Res. 2002;21:181–205. doi: 10.1080/09712119.2002.9706369. - DOI
-
- Van Soest PJ. Nutritional Ecology of the Ruminant. Cornell University Press; 1994.
-
- Gensa U. Review on cyanide poisoning in ruminants. J. Biol. Agric. Healthc. 2019;6:1–12.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

