Review of fragmentation of synthetic single-stranded oligonucleotides by tandem mass spectrometry from 2014 to 2022
- PMID: 37580500
- PMCID: PMC10909466
- DOI: 10.1002/rcm.9596
Review of fragmentation of synthetic single-stranded oligonucleotides by tandem mass spectrometry from 2014 to 2022
Abstract
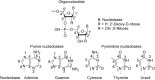
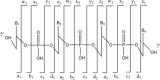
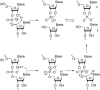
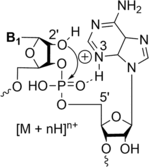
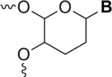
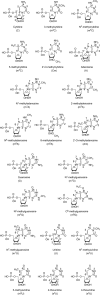
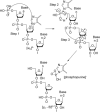
The fragmentation of oligonucleotides by mass spectrometry allows for the determination of their sequences. It is necessary to understand how oligonucleotides dissociate in the gas phase, which allows interpretation of data to obtain sequence information. Since 2014, a range of fragmentation mechanisms, including a novel internal rearrangement, have been proposed using different ion dissociation techniques. The recent publications have focused on the fragmentation of modified oligonucleotides such as locked nucleic acids, modified nucleobases (methylated, spacer, nebularine and aminopurine) and modification to the carbon 2'-position on the sugar ring; these modified oligonucleotides are of great interest as therapeutics. Comparisons of different dissociation techniques have been reported, including novel approaches such as plasma electron detachment dissociation and radical transfer dissociation. This review covers the period 2014-2022 and details the new knowledge gained with respect to oligonucleotide dissociation using tandem mass spectrometry (without priori sample digestion) during that time, with a specific focus on synthetic single-stranded oligonucleotides.
© 2023 The Authors. Rapid Communications in Mass Spectrometry published by John Wiley & Sons Ltd.
Figures







Similar articles
-
Electron transfer followed by collision-induced dissociation (NET-CID) for generating sequence information from backbone-modified oligonucleotide anions.Rapid Commun Mass Spectrom. 2013 Jan 15;27(1):249-57. doi: 10.1002/rcm.6428. Rapid Commun Mass Spectrom. 2013. PMID: 23239339
-
Fast Electron Detachment Dissociation of Oligonucleotides in Electron-Nitrogen Plasma Stored in Magneto Radio-Frequency Ion Traps.Anal Chem. 2022 Nov 8;94(44):15510-15517. doi: 10.1021/acs.analchem.2c04027. Epub 2022 Oct 24. Anal Chem. 2022. PMID: 36279405
-
Investigation of the Influence of Charge State and Collision Energy on Oligonucleotide Fragmentation by Tandem Mass Spectrometry.Molecules. 2023 Jan 25;28(3):1169. doi: 10.3390/molecules28031169. Molecules. 2023. PMID: 36770836 Free PMC article.
-
Characterization of nucleic acids by tandem mass spectrometry - The second decade (2004-2013): From DNA to RNA and modified sequences.Mass Spectrom Rev. 2016 Jul;35(4):483-523. doi: 10.1002/mas.21442. Epub 2014 Oct 6. Mass Spectrom Rev. 2016. PMID: 25288464 Review.
-
Elucidation of nucleic acid-drug interactions by tandem mass spectrometry.Chimia (Aarau). 2014;68(3):164-7. doi: 10.2533/chimia.2014.164. Chimia (Aarau). 2014. PMID: 24801849 Review.
Cited by
-
Mass Spectrometry Strategies for O-Glycoproteomics.Cells. 2024 Feb 25;13(5):394. doi: 10.3390/cells13050394. Cells. 2024. PMID: 38474358 Free PMC article. Review.
-
Distinguishing Isomeric Cyclobutane Thymidine Dimers by Ion Mobility and Tandem Mass Spectrometry.J Am Soc Mass Spectrom. 2024 Aug 7;35(8):1768-1774. doi: 10.1021/jasms.4c00133. Epub 2024 Jul 2. J Am Soc Mass Spectrom. 2024. PMID: 38952267 Free PMC article.
-
Non-Targeted Detection of Synthetic Oligonucleotides in Equine Serum Using Liquid Chromatography-High-Resolution Mass Spectrometry.Int J Mol Sci. 2024 May 25;25(11):5752. doi: 10.3390/ijms25115752. Int J Mol Sci. 2024. PMID: 38891955 Free PMC article.
-
Ion Mobility Gas-Phase Separation Enhances Top-Down Mass Spectrometry of Heavily Modified Guide RNA.Anal Chem. 2025 May 6;97(17):9430-9437. doi: 10.1021/acs.analchem.5c00705. Epub 2025 Apr 11. Anal Chem. 2025. PMID: 40215333 Free PMC article.
-
Sequencing of Phosphorodiamidate Morpholino Oligomers by Hydrophilic Interaction Chromatography Coupled to Tandem Mass Spectrometry.J Am Soc Mass Spectrom. 2024 Oct 2;35(10):2490-2498. doi: 10.1021/jasms.4c00281. Epub 2024 Aug 30. J Am Soc Mass Spectrom. 2024. PMID: 39213635 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
