Metabolic profiling stratifies colorectal cancer and reveals adenosylhomocysteinase as a therapeutic target
- PMID: 37580540
- PMCID: PMC10447251
- DOI: 10.1038/s42255-023-00857-0
Metabolic profiling stratifies colorectal cancer and reveals adenosylhomocysteinase as a therapeutic target
Abstract
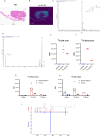
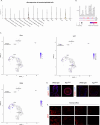
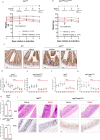
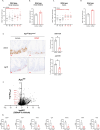
The genomic landscape of colorectal cancer (CRC) is shaped by inactivating mutations in tumour suppressors such as APC, and oncogenic mutations such as mutant KRAS. Here we used genetically engineered mouse models, and multimodal mass spectrometry-based metabolomics to study the impact of common genetic drivers of CRC on the metabolic landscape of the intestine. We show that untargeted metabolic profiling can be applied to stratify intestinal tissues according to underlying genetic alterations, and use mass spectrometry imaging to identify tumour, stromal and normal adjacent tissues. By identifying ions that drive variation between normal and transformed tissues, we found dysregulation of the methionine cycle to be a hallmark of APC-deficient CRC. Loss of Apc in the mouse intestine was found to be sufficient to drive expression of one of its enzymes, adenosylhomocysteinase (AHCY), which was also found to be transcriptionally upregulated in human CRC. Targeting of AHCY function impaired growth of APC-deficient organoids in vitro, and prevented the characteristic hyperproliferative/crypt progenitor phenotype driven by acute deletion of Apc in vivo, even in the context of mutant Kras. Finally, pharmacological inhibition of AHCY reduced intestinal tumour burden in ApcMin/+ mice indicating its potential as a metabolic drug target in CRC.
© 2023. The Author(s).
Conflict of interest statement
O.D.K.M. is a co-founder, shareholder and board member of Faeth Therapeutics. The remaining authors declare no competing interests.
Figures









References
-
- Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). - PubMed
-
- Balog, J. et al. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci. Transl. Med.5, 194ra193 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

