Single-cell analysis of human MAIT cell transcriptional, functional and clonal diversity
- PMID: 37580605
- PMCID: PMC10457204
- DOI: 10.1038/s41590-023-01575-1
Single-cell analysis of human MAIT cell transcriptional, functional and clonal diversity
Abstract
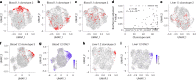
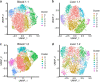
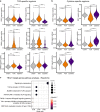
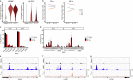
Mucosal-associated invariant T (MAIT) cells are innate-like T cells that recognize microbial metabolites through a semi-invariant T cell receptor (TCR). Major questions remain regarding the extent of human MAIT cell functional and clonal diversity. To address these, we analyzed the single-cell transcriptome and TCR repertoire of blood and liver MAIT cells and developed functional RNA-sequencing, a method to integrate function and TCR clonotype at single-cell resolution. MAIT cell clonal diversity was comparable to conventional memory T cells, with private TCR repertoires shared across matched tissues. Baseline functional diversity was low and largely related to tissue site. MAIT cells showed stimulus-specific transcriptional responses in vitro, with cells positioned along gradients of activation. Clonal identity influenced resting and activated transcriptional profiles but intriguingly was not associated with the capacity to produce IL-17. Overall, MAIT cells show phenotypic and functional diversity according to tissue localization, stimulation environment and clonotype.
© 2023. The Author(s).
Conflict of interest statement
P.K. has acted as a consultant to UCB, Biomunex, AstraZeneca and Infinitopes. N.M.P. has acted as a consultant to Infinitopes. The other authors declare no competing interests.
Figures














References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

