The RNA-binding protein KSRP aggravates malignant progression of clear cell renal cell carcinoma through transcriptional inhibition and post-transcriptional destabilization of the NEDD4L ubiquitin ligase
- PMID: 37580757
- PMCID: PMC10424398
- DOI: 10.1186/s12929-023-00949-9
The RNA-binding protein KSRP aggravates malignant progression of clear cell renal cell carcinoma through transcriptional inhibition and post-transcriptional destabilization of the NEDD4L ubiquitin ligase
Abstract
Background: KH-type splicing regulatory protein (KHSRP, also called KSRP), a versatile RNA-binding protein, plays a critical role in various physiological and pathological conditions through modulating gene expressions at multiple levels. However, the role of KSRP in clear cell renal cell carcinoma (ccRCC) remains poorly understood.
Methods: KSRP expression was detected by a ccRCC tissue microarray and evaluated by an in silico analysis. Cell loss-of-function and gain-of-function, colony-formation, anoikis, and transwell assays, and an orthotopic bioluminescent xenograft model were conducted to determine the functional role of KRSP in ccRCC progression. Micro (mi)RNA and complementary (c)DNA microarrays were used to identify downstream targets of KSRP. Western blotting, quantitative real-time polymerase chain reaction, and promoter- and 3-untranslated region (3'UTR)-luciferase reporter assays were employed to validate the underlying mechanisms of KSRP which aggravate progression of ccRCC.
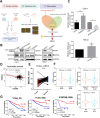
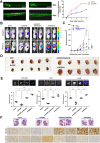
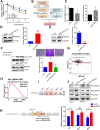
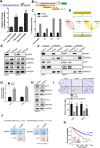
Results: Our results showed that dysregulated high levels of KSRP were correlated with advanced clinical stages, larger tumor sizes, recurrence, and poor prognoses of ccRCC. Neural precursor cell-expressed developmentally downregulated 4 like (NEDD4L) was identified as a novel target of KSRP, which can reverse the protumorigenic and prometastatic characteristics as well as epithelial-mesenchymal transition (EMT) promotion by KSRP in vitro and in vivo. Molecular studies revealed that KSRP can decrease NEDD4L messenger (m)RNA stability via inducing mir-629-5p upregulation and directly targeting the AU-rich elements (AREs) of the 3'UTR. Moreover, KSRP was shown to transcriptionally suppress NEDD4L via inducing the transcriptional repressor, Wilm's tumor 1 (WT1). In the clinic, ccRCC samples revealed a positive correlation between KSRP and mesenchymal-related genes, and patients expressing high KSRP and low NEDD4L had the worst prognoses.
Conclusion: The current findings unveil novel mechanisms of KSRP which promote malignant progression of ccRCC through transcriptional inhibition and post-transcriptional destabilization of NEDD4L transcripts. Targeting KSRP and its pathways may be a novel pharmaceutical intervention for ccRCC.
Keywords: EMT; KSRP; NEDD4L; Progression; ccRCC.
© 2023. National Science Council of the Republic of China (Taiwan).
Conflict of interest statement
The authors declare that no competing interests exist.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

