A novel immune modulator IM33 mediates a glia-gut-neuronal axis that controls lifespan
- PMID: 37582366
- PMCID: PMC10592285
- DOI: 10.1016/j.neuron.2023.07.010
A novel immune modulator IM33 mediates a glia-gut-neuronal axis that controls lifespan
Abstract
Aging is a complex process involving various systems and behavioral changes. Altered immune regulation, dysbiosis, oxidative stress, and sleep decline are common features of aging, but their interconnection is poorly understood. Using Drosophila, we discover that IM33, a novel immune modulator, and its mammalian homolog, secretory leukocyte protease inhibitor (SLPI), are upregulated in old flies and old mice, respectively. Knockdown of IM33 in glia elevates the gut reactive oxygen species (ROS) level and alters gut microbiota composition, including increased Lactiplantibacillus plantarum abundance, leading to a shortened lifespan. Additionally, dysbiosis induces sleep fragmentation through the activation of insulin-producing cells in the brain, which is mediated by the binding of Lactiplantibacillus plantarum-produced DAP-type peptidoglycan to the peptidoglycan recognition protein LE (PGRP-LE) receptor. Therefore, IM33 plays a role in the glia-microbiota-neuronal axis, connecting neuroinflammation, dysbiosis, and sleep decline during aging. Identifying molecular mediators of these processes could lead to the development of innovative strategies for extending lifespan.
Keywords: Drosophila; IM33; brain-gut axis; glia-gut axis; longevity; neuroimmunology.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



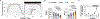
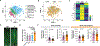

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

