Spatial transcriptomics reveal markers of histopathological changes in Duchenne muscular dystrophy mouse models
- PMID: 37582915
- PMCID: PMC10427630
- DOI: 10.1038/s41467-023-40555-9
Spatial transcriptomics reveal markers of histopathological changes in Duchenne muscular dystrophy mouse models
Abstract
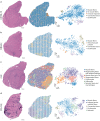
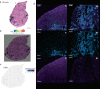
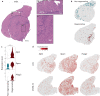
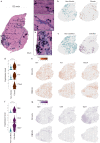
Duchenne muscular dystrophy is caused by mutations in the DMD gene, leading to lack of dystrophin. Chronic muscle damage eventually leads to histological alterations in skeletal muscles. The identification of genes and cell types driving tissue remodeling is a key step to developing effective therapies. Here we use spatial transcriptomics in two Duchenne muscular dystrophy mouse models differing in disease severity to identify gene expression signatures underlying skeletal muscle pathology and to directly link gene expression to muscle histology. We perform deconvolution analysis to identify cell types contributing to histological alterations. We show increased expression of specific genes in areas of muscle regeneration (Myl4, Sparc, Hspg2), fibrosis (Vim, Fn1, Thbs4) and calcification (Bgn, Ctsk, Spp1). These findings are confirmed by smFISH. Finally, we use differentiation dynamic analysis in the D2-mdx muscle to identify muscle fibers in the present state that are predicted to become affected in the future state.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Mendell JR, Lloyd-Puryear M. Report Of MDA Muscle Disease Symposium On Newborn Screening For Duchenne Muscular Dystrophy. Muscle Nerve. 2013;48:21–26. - PubMed
-
- Hoffman EP, Brown RH, Kunkel LM. Dystrophin: The protein product of the duchenne muscular dystrophy locus. Cell. 1987;51:919–928. - PubMed
-
- Le Rumeur E, Winder SJ, Hubert JF. Dystrophin: More than just the sum of its parts. Biochim. et. Biophys. Acta - Proteins Proteom. 2010;1804:1713–1722. - PubMed
-
- Ervasti JM. Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim. et. Biophys. Acta - Mol. Basis Dis. 2007;1772:108–117. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

