AAV-mediated base-editing therapy ameliorates the disease phenotypes in a mouse model of retinitis pigmentosa
- PMID: 37582961
- PMCID: PMC10427680
- DOI: 10.1038/s41467-023-40655-6
AAV-mediated base-editing therapy ameliorates the disease phenotypes in a mouse model of retinitis pigmentosa
Abstract
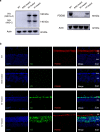
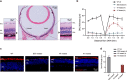
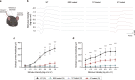
Base editing technology is an ideal solution for treating pathogenic single-nucleotide variations (SNVs). No gene editing therapy has yet been approved for eye diseases, such as retinitis pigmentosa (RP). Here, we show, in the rd10 mouse model, which carries an SNV identified as an RP-causing mutation in human patients, that subretinal delivery of an optimized dual adeno-associated virus system containing the adenine base editor corrects the pathogenic SNV in the neuroretina with up to 49% efficiency. Light microscopy showed that a thick and robust outer nuclear layer (photoreceptors) was preserved in the treated area compared with the thin, degenerated outer nuclear layer without treatment. Substantial electroretinogram signals were detected in treated rd10 eyes, whereas control treated eyes showed minimal signals. The water maze experiment showed that the treatment substantially improved vision-guided behavior. Together, we construct and validate a translational therapeutic solution for the treatment of RP in humans. Our findings might accelerate the development of base-editing based gene therapies.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

