Identification of novel gene signature for lung adenocarcinoma by machine learning to predict immunotherapy and prognosis
- PMID: 37583701
- PMCID: PMC10424935
- DOI: 10.3389/fimmu.2023.1177847
Identification of novel gene signature for lung adenocarcinoma by machine learning to predict immunotherapy and prognosis
Abstract
Background: Lung adenocarcinoma (LUAD) as a frequent type of lung cancer has a 5-year overall survival rate of lower than 20% among patients with advanced lung cancer. This study aims to construct a risk model to guide immunotherapy in LUAD patients effectively.
Materials and methods: LUAD Bulk RNA-seq data for the construction of a model, single-cell RNA sequencing (scRNA-seq) data (GSE203360) for cell cluster analysis, and microarray data (GSE31210) for validation were collected from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) database. We used the Seurat R package to filter and process scRNA-seq data. Sample clustering was performed in the ConsensusClusterPlus R package. Differentially expressed genes (DEGs) between two groups were mined by the Limma R package. MCP-counter, CIBERSORT, ssGSEA, and ESTIMATE were employed to evaluate immune characteristics. Stepwise multivariate analysis, Univariate Cox analysis, and Lasso regression analysis were conducted to identify key prognostic genes and were used to construct the risk model. Key prognostic gene expressions were explored by RT-qPCR and Western blot assay.
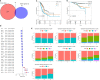
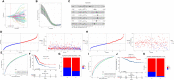
Results: A total of 27 immune cell marker genes associated with prognosis were identified for subtyping LUAD samples into clusters C3, C2, and C1. C1 had the longest overall survival and highest immune infiltration among them, followed by C2 and C3. Oncogenic pathways such as VEGF, EFGR, and MAPK were more activated in C3 compared to the other two clusters. Based on the DEGs among clusters, we confirmed seven key prognostic genes including CPA3, S100P, PTTG1, LOXL2, MELTF, PKP2, and TMPRSS11E. Two risk groups defined by the seven-gene risk model presented distinct responses to immunotherapy and chemotherapy, immune infiltration, and prognosis. The mRNA and protein level of CPA3 was decreased, while the remaining six gene levels were increased in clinical tumor tissues.
Conclusion: Immune cell markers are effective in clustering LUAD samples into different subtypes, and they play important roles in regulating the immune microenvironment and cancer development. In addition, the seven-gene risk model may serve as a guide for assisting in personalized treatment in LUAD patients.
Keywords: immune cells; immunotherapy; lung adenocarcinoma; molecular subtyping; risk model; single-cell analysis.
Copyright © 2023 Shu, Jiang and Zhao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures









References
-
- Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet (2018) 391(10125):1023–75. doi: 10.1016/S0140-6736(17)33326-3 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

