Molecular dynamics and simulation analysis against superoxide dismutase (SOD) target of Micrococcus luteus with secondary metabolites from Bacillus licheniformis recognized by genome mining approach
- PMID: 37583871
- PMCID: PMC10424208
- DOI: 10.1016/j.sjbs.2023.103753
Molecular dynamics and simulation analysis against superoxide dismutase (SOD) target of Micrococcus luteus with secondary metabolites from Bacillus licheniformis recognized by genome mining approach
Abstract
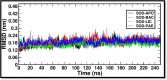
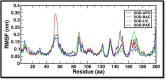
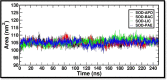
Micrococcus luteus, also known as M. luteus, is a bacterium that inhabits mucous membranes, human skin, and various environmental sources. It is commonly linked to infections, especially among individuals who have compromised immune systems. M. luteus is capable of synthesizing the enzyme superoxide dismutase (SOD) as a component of its protective response to reactive oxygen species (ROS). This enzyme serves as a promising target for drug development in various diseases. The current study utilized a subtractive genomics approach to identify potential therapeutic targets from M. luteus. Additionally, genome mining was employed to identify and characterize the biosynthetic gene clusters (BGCs) responsible for the production of secondary metabolites in Bacillus licheniformis (B. licheniformis), a bacterium known for its production of therapeutically relevant secondary metabolites. Subtractive genomics resulted in identification of important extracellular protein SOD as a drug target that plays a crucial role in shielding cells from damage caused by ROS. Genome mining resulted in identification of five potential ligands (secondary metabolites) from B. licheniformis such as, Bacillibactin (BAC), Paenibactin (PAE), Fengycin (FEN), Surfactin (SUR) and Lichenysin (LIC). Molecular docking was used to predict and analyze the binding interactions between these five ligands and target protein SOD. The resulting protein-ligand complexes were further analyzed for their motions and interactions of atoms and molecules over 250 ns using molecular dynamics (MD) simulation analysis. The analysis of MD simulations suggests, Bacillibactin as the probable candidate to arrest the activities of SOD. All the five compounds reported in this study were found to act by directly/indirectly interacting with ROS molecules, such as superoxide radicals (O2-) and hydrogen peroxide (H2O2), and transforming them into less reactive species. This antioxidant activity contributes to its protective effects against oxidative stress-induced damage in cells making them likely candidate for various applications, including in the development of antioxidant-based therapies, nutraceuticals, and functional foods.
Keywords: Bacillus licheniformis; Genome mining; Micrococcus luteus; Molecular dynamics; Simulation.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Gruning B.A., Guerler A., Hillman-Jackson J., Hiltemann S., Jalili V., Rasche H., Soranzo N., Goecks J., Taylor J., Nekrutenko A., Blankenberg D. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. - DOI - PMC - PubMed
-
- Aly, R., 1991. Cutaneous microbiology. Dermatology. Los Altos: Appleton & Lange. 22-25.
-
- Bagewadi Z.K., Aakanksha U.K., Yaraguppi D.A., Yunus Khan T.M., Deshpande S.H., Dammalli M., Revankar A.G., Savalagi A.J., Hiremath S.V. Molecular docking and simulation studies against nucleoside diphosphate kinase (ndk) of pseudomonas aeruginosa with secondary metabolite identified by genome mining from paenibacillusehimensis. J. Biomol. Struct. Dyn. 2023:1–10. doi: 10.1080/07391102.2023.2167118. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

