Anomaly detection in mixed high-dimensional molecular data
- PMID: 37584673
- PMCID: PMC10457663
- DOI: 10.1093/bioinformatics/btad501
Anomaly detection in mixed high-dimensional molecular data
Abstract
Motivation: Mixed molecular data combines continuous and categorical features of the same samples, such as OMICS profiles with genotypes, diagnoses, or patient sex. Like all high-dimensional molecular data, it is prone to incorrect values that can stem from various sources for example the technical limitations of the measurement devices, errors in the sample preparation, or contamination. Most anomaly detection algorithms identify complete samples as outliers or anomalies. However, in most cases, not all measurements of those samples are erroneous but only a few one-dimensional features within the samples are incorrect. These one-dimensional data errors are continuous measurements that are either located outside or inside the normal ranges of their features but in both cases show atypical values given all other continuous and categorical features in the sample. Additionally, categorical anomalies can occur for example when the genotype or diagnosis was submitted wrongly.
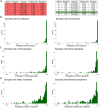
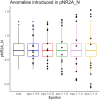
Results: We introduce ADMIRE (Anomaly Detection using MIxed gRaphical modEls), a novel approach for the detection and correction of anomalies in mixed high-dimensional data. Hereby, we focus on the detection of single (one-dimensional) data errors in the categorical and continuous features of a sample. For that the joint distribution of continuous and categorical features is learned by mixed graphical models, anomalies are detected by the difference between measured and model-based estimations and are corrected using imputation. We evaluated ADMIRE in simulation and by screening for anomalies in one of our own metabolic datasets. In simulation experiments, ADMIRE outperformed the state-of-the-art methods of Local Outlier Factor, stray, and Isolation Forest.
Availability and implementation: All data and code is available at https://github.com/spang-lab/adadmire. ADMIRE is implemented in a Python package called adadmire which can be found at https://pypi.org/project/adadmire.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures



References
-
- Ando S. Clustering needles in a haystack: an information theoretic analysis of minority and outlier detection. In: Seventh IEEE International Conference on Data Mining (ICDM 2007), btad501. IEEE 2007, pp. 13–22.
-
- Breunig MM, Kriegel H-P, Ng RT. et al. Lof. SIGMOD Rec 2000;29:93–104.
-
- Cheng J, Li T, Levina E. et al. High-dimensional mixed graphical models. J Comput Graph Stat 2017;26:367–78.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

