Emergence of a new designated clade 16 with significant antigenic drift in hemagglutinin gene of H9N2 subtype avian influenza virus in eastern China
- PMID: 37585307
- PMCID: PMC10467529
- DOI: 10.1080/22221751.2023.2249558
Emergence of a new designated clade 16 with significant antigenic drift in hemagglutinin gene of H9N2 subtype avian influenza virus in eastern China
Abstract
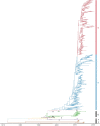
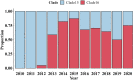
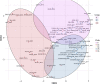
H9N2 avian influenza viruses (AIVs) pose an increasing threat to the poultry industry worldwide and have pandemic potential. Vaccination has been principal prevention strategy to control H9N2 in China since 1998, but vaccine effectiveness is persistently challenged by the emergence of the genetic and/or antigenic variants. Here, we analysed the genetic and antigenic characteristics of H9N2 viruses in China, including 70 HA sequences of H9N2 isolates from poultry, 7358 from online databases during 2010-2020, and 15 from the early reference strains. Bayesian analyses based on hemagglutinin (HA) gene revealed that a new designated clade16 emerged in April 2012, and was prevalent and co-circulated with clade 15 since 2013 in China. Clade 16 viruses exhibited decreased cross-reactivity with those from clade 15. Antigenic Cartography analyses showed represent strains were classified into three antigenic groups named as Group1, Group2 and Group3, and most of the strains in Group 3 (15/17, 88.2%) were from Clade 16 while most of the strains in Group2 (26/29, 89.7%) were from Clade 15. The mean distance between Group 3 and Group 2 was 4.079 (95%CI 3.605-4.554), revealing that major switches to antigenic properties were observed over the emergence of clade 16. Genetic analysis indicated that 11 coevolving amino acid substitutions primarily at antigenic sites were associated with the antigenic differences between clade 15 and clade 16. These data highlight complexities of the genetic evolution and provide a framework for the genetic basis and antigenic characterization of emerging clade 16 of H9N2 subtype avian influenza virus.
Keywords: H9N2; HA gene; antigenic; genetic evolution; new designated clade.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
References
-
- Chen PL, Zhang ZJ.. Isolation and serological identification of avian influenza A virus from chickens. Chinese Poultry. 1997;11:4–6.
-
- Guo J, Wang Y, Zhao C, et al. . Molecular characterization, receptor binding property, and replication in chickens and mice of H9N2 avian influenza viruses isolated from chickens, peafowls, and wild birds in eastern China. Emerg Microbes Infect. 2021;10(1):2098–2112. DOI:10.1080/22221751.2021.1999778 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
