Genetic diversity, phylogenetic and phylogeographic analysis of Anopheles culicifacies species complex using ITS2 and COI sequences
- PMID: 37585421
- PMCID: PMC10431676
- DOI: 10.1371/journal.pone.0290178
Genetic diversity, phylogenetic and phylogeographic analysis of Anopheles culicifacies species complex using ITS2 and COI sequences
Abstract
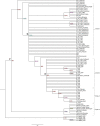
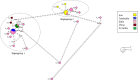
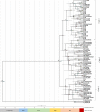
Anopheles culicifacies is the major vector of malaria in Sri Lanka and the Indian subcontinent which is characterized as a species complex with five sibling species provisionally designated as A, B, C, D and E. The current study was carried out to understand the phylogenetic and phylogeographic relationships between the sibling species of the species complex while observing their genetic diversity and genetic differentiation. Thirty-five ITS2 and seventy-seven COI sequences of An. culicifacies species complex reported from different geographical locations of Asia and China at the NCBI public database were used for the analysis. Bayesian likelihood trees were generated for the phylogenetic analysis. The divergence of the species complex was obtained from the Bayesian phylogeographic model in BEAST. There were two clades of the sibling species of An. culicifacies species complex as A, D and B, C and E in both phylogenetic and phylogeographic analysis using ITS2 sequences. Based on the highly divergent COI sequences and the high mutation rate of the mitochondrial genome, there were four and three clades in both phylogenetic and phylogeographic analysis using COI sequences. The diversification of An. culicifacies species complex was obtained as ranging from 20.25 to 24.12 Mya and 22.37 to 26.22 Mya based on ITS2 and COI phylogeographic analysis respectively. There was a recent diversification of the sibling species A and D than the sibling species B, C and E. Low haplotype diversity was observed in the sequences reported from Sri Lanka in both ITS2 and COI analysis that can be due to bottlenecks resulting from the intense malaria control efforts. A high genetic differentiation was achieved for some populations due to the large geographical distance. The high genetic diversity based on the five sibling species implies the possibility of maintaining a relatively high effective population size despite the vector control efforts.
Copyright: © 2023 Rathnayake et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Surendran SN, Abhayawardana TA, De Silva BGDNK, Ramasamy R, Ramasamy MS . Anopheles culicifacies Y-chromosome dimorphism indicates sibling species (b and e) with different malaria vector potential in Sri Lanka. Medical and Veterinary Entomology. 2000; 14(4): 437–440. doi: 10.1046/j.1365-2915.2000.00264.x . - DOI - PubMed
-
- Shanika KHS, Harischandra IN, De Silva BGDNK. Nucleotide sequence and secondary structure variations in ITS2-rDNA region of the members of Anopheles culicifacies (Diptera: Culicidae) species complex Nucleotide Sequence and Secondary Structure Variations in ITS2-rDNA Region of the Members of Anopheles culicifacies (Diptera: Culicidae) Species Complex. Vidyodaya Journal of Science. 2016; 20: 26–40.
-
- Goswami G, Raghavendra K, Nanda N, Gakhar SK, Subbarao SK. PCR-RFLP of mitochondrial cytochrome oxidase subunit II and ITS2 of ribosomal DNA: Markers for the identification of members of the Anopheles culicifacies complex (Diptera: Culicidae). Acta Tropica. 2005; 95(2): 92–99. doi: 10.1016/j.actatropica.2005.04.018 . - DOI - PubMed
-
- Van Bortel W, Sochanta T, Harbach RE, Socheat D, Roelants P, Backeljau T, et al.. Presence of Anopheles culicifacies B in Cambodia established by the PCR-RFLP assay developed for the identification of Anopheles minimus species A and C and four related species. Medical and Veterinary Entomology. 2002; 16(3): 329–334. doi: 10.1046/j.1365-2915.2002.00376.x . - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

