A TGF-β/KLF10 signaling axis regulates atrophy-associated genes to induce muscle wasting in pancreatic cancer
- PMID: 37585460
- PMCID: PMC10462925
- DOI: 10.1073/pnas.2215095120
A TGF-β/KLF10 signaling axis regulates atrophy-associated genes to induce muscle wasting in pancreatic cancer
Abstract
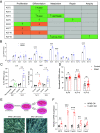
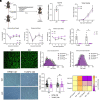
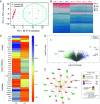
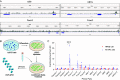
Cancer cachexia, and its associated complications, represent a large and currently untreatable roadblock to effective cancer management. Many potential therapies have been proposed and tested-including appetite stimulants, targeted cytokine blockers, and nutritional supplementation-yet highly effective therapies are lacking. Innovative approaches to treating cancer cachexia are needed. Members of the Kruppel-like factor (KLF) family play wide-ranging and important roles in the development, maintenance, and metabolism of skeletal muscle. Within the KLF family, we identified KLF10 upregulation in a multitude of wasting contexts-including in pancreatic, lung, and colon cancer mouse models as well as in human patients. We subsequently interrogated loss-of-function of KLF10 as a potential strategy to mitigate cancer associated muscle wasting. In vivo studies leveraging orthotopic implantation of pancreas cancer cells into wild-type and KLF10 KO mice revealed significant preservation of lean mass and robust suppression of pro-atrophy muscle-specific ubiquitin ligases Trim63 and Fbxo32, as well as other factors implicated in atrophy, calcium signaling, and autophagy. Bioinformatics analyses identified Transforming growth factor beta (TGF-β), a known inducer of KLF10 and cachexia promoting factor, as a key upstream regulator of KLF10. We provide direct in vivo evidence that KLF10 KO mice are resistant to the atrophic effects of TGF-β. ChIP-based binding studies demonstrated direct binding to Trim63, a known wasting-associated atrogene. Taken together, we report a critical role for the TGF-β/KLF10 axis in the etiology of pancreatic cancer-associated muscle wasting and highlight the utility of targeting KLF10 as a strategy to prevent muscle wasting and limit cancer-associated cachexia.
Keywords: KLF10; TGF-β; cachexia; muscle wasting; pancreatic cancer.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Dewys W. D., et al. , Prognostic effect of weight loss prior to chemotherapy in cancer patients. Eastern Cooperative Oncology Group. Am. J. Med. 69, 491–497 (1980). - PubMed
-
- Bachmann J., et al. , Cachexia worsens prognosis in patients with resectable pancreatic cancer. J. Gastrointest. Surg. 12, 1193–1201 (2008). - PubMed
-
- Baracos V. E., Martin L., Korc M., Guttridge D. C., Fearon K. C. H., Cancer-associated cachexia. Nat. Rev. Dis. Primers 4, 17105 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

