Loss of function of the ALS-associated NEK1 kinase disrupts microtubule homeostasis and nuclear import
- PMID: 37585529
- PMCID: PMC10431718
- DOI: 10.1126/sciadv.adi5548
Loss of function of the ALS-associated NEK1 kinase disrupts microtubule homeostasis and nuclear import
Abstract
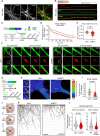
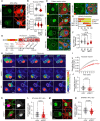
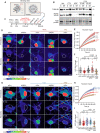
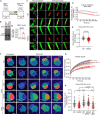
Loss-of-function variants in NIMA-related kinase 1 (NEK1) constitute a major genetic cause of amyotrophic lateral sclerosis (ALS), accounting for 2 to 3% of all cases. However, how NEK1 mutations cause motor neuron (MN) dysfunction is unknown. Using mass spectrometry analyses for NEK1 interactors and NEK1-dependent expression changes, we find functional enrichment for proteins involved in the microtubule cytoskeleton and nucleocytoplasmic transport. We show that α-tubulin and importin-β1, two key proteins involved in these processes, are phosphorylated by NEK1 in vitro. NEK1 is essential for motor control and survival in Drosophila models in vivo, while using several induced pluripotent stem cell (iPSC)-MN models, including NEK1 knockdown, kinase inhibition, and a patient mutation, we find evidence for disruptions in microtubule homeostasis and nuclear import. Notably, stabilizing microtubules with two distinct classes of drugs restored NEK1-dependent deficits in both pathways. The capacity of NEK1 to modulate these processes that are critically involved in ALS pathophysiology renders this kinase a formidable therapeutic candidate.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

