RNA polymerase II pausing temporally coordinates cell cycle progression and erythroid differentiation
- PMID: 37586368
- PMCID: PMC10615711
- DOI: 10.1016/j.devcel.2023.07.018
RNA polymerase II pausing temporally coordinates cell cycle progression and erythroid differentiation
Abstract
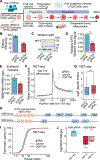
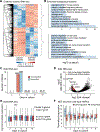
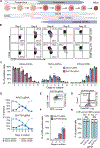
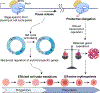
Controlled release of promoter-proximal paused RNA polymerase II (RNA Pol II) is crucial for gene regulation. However, studying RNA Pol II pausing is challenging, as pause-release factors are almost all essential. In this study, we identified heterozygous loss-of-function mutations in SUPT5H, which encodes SPT5, in individuals with β-thalassemia. During erythropoiesis in healthy human cells, cell cycle genes were highly paused as cells transition from progenitors to precursors. When the pathogenic mutations were recapitulated by SUPT5H editing, RNA Pol II pause release was globally disrupted, and as cells began transitioning from progenitors to precursors, differentiation was delayed, accompanied by a transient lag in erythroid-specific gene expression and cell cycle kinetics. Despite this delay, cells terminally differentiate, and cell cycle phase distributions normalize. Therefore, hindering pause release perturbs proliferation and differentiation dynamics at a key transition during erythropoiesis, identifying a role for RNA Pol II pausing in temporally coordinating the cell cycle and erythroid differentiation.
Keywords: RNA Pol II pausing; SPT5; SUPT5H; cell cycle; differentiation; erythropoiesis; hemoglobin; human genetic variation; transcriptional regulation.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests V.G.S. serves as an advisor to and/or has equity in Branch Biosciences, Ensoma, Novartis, Forma, Sana Biotechnology, and Cellarity, all unrelated to the present work. J.C.U. is an employee of Illumina, Inc., unrelated to the present work. R.I. is a founder, board member, and shareholder of Cellforma, unrelated to the present work.
Figures



Update of
-
RNA Polymerase II pausing temporally coordinates cell cycle progression and erythroid differentiation.medRxiv [Preprint]. 2023 Mar 7:2023.03.03.23286760. doi: 10.1101/2023.03.03.23286760. medRxiv. 2023. Update in: Dev Cell. 2023 Oct 23;58(20):2112-2127.e4. doi: 10.1016/j.devcel.2023.07.018. PMID: 36945604 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

