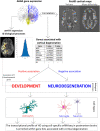
Genetic topography and cortical cell loss in Huntington's disease link development and neurodegeneration
- PMID: 37587097
- PMCID: PMC10629790
- DOI: 10.1093/brain/awad275
Genetic topography and cortical cell loss in Huntington's disease link development and neurodegeneration
Abstract
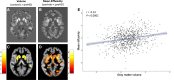
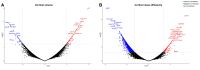
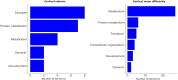
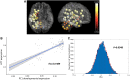
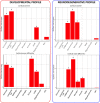
Cortical cell loss is a core feature of Huntington's disease (HD), beginning many years before clinical motor diagnosis, during the premanifest stage. However, it is unclear how genetic topography relates to cortical cell loss. Here, we explore the biological processes and cell types underlying this relationship and validate these using cell-specific post-mortem data. Eighty premanifest participants on average 15 years from disease onset and 71 controls were included. Using volumetric and diffusion MRI we extracted HD-specific whole brain maps where lower grey matter volume and higher grey matter mean diffusivity, relative to controls, were used as proxies of cortical cell loss. These maps were combined with gene expression data from the Allen Human Brain Atlas (AHBA) to investigate the biological processes relating genetic topography and cortical cell loss. Cortical cell loss was positively correlated with the expression of developmental genes (i.e. higher expression correlated with greater atrophy and increased diffusivity) and negatively correlated with the expression of synaptic and metabolic genes that have been implicated in neurodegeneration. These findings were consistent for diffusion MRI and volumetric HD-specific brain maps. As wild-type huntingtin is known to play a role in neurodevelopment, we explored the association between wild-type huntingtin (HTT) expression and developmental gene expression across the AHBA. Co-expression network analyses in 134 human brains free of neurodegenerative disorders were also performed. HTT expression was correlated with the expression of genes involved in neurodevelopment while co-expression network analyses also revealed that HTT expression was associated with developmental biological processes. Expression weighted cell-type enrichment (EWCE) analyses were used to explore which specific cell types were associated with HD cortical cell loss and these associations were validated using cell specific single nucleus RNAseq (snRNAseq) data from post-mortem HD brains. The developmental transcriptomic profile of cortical cell loss in preHD was enriched in astrocytes and endothelial cells, while the neurodegenerative transcriptomic profile was enriched for neuronal and microglial cells. Astrocyte-specific genes differentially expressed in HD post-mortem brains relative to controls using snRNAseq were enriched in the developmental transcriptomic profile, while neuronal and microglial-specific genes were enriched in the neurodegenerative transcriptomic profile. Our findings suggest that cortical cell loss in preHD may arise from dual pathological processes, emerging as a consequence of neurodevelopmental changes, at the beginning of life, followed by neurodegeneration in adulthood, targeting areas with reduced expression of synaptic and metabolic genes. These events result in age-related cell death across multiple brain cell types.
Keywords: Huntington’s disease; atrophy; diffusivity; gene expression; imaging transcriptomics.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
S.J.T was the global principal investigator (PI) for TrackOn-HD. A.D., B.R.L., R.A.C.R. and B.L. were site PIs for Paris, Vancouver, Leiden and Ulm, respectively. S.J.T. receives research grant funding from the CHDI Foundation, Vertex Pharmaceuticals, the UK Medical Research Council, the Wellcome Trust (200181/Z/15/Z), and the UK Dementia Research Institute that receives its funding from DRI Ltd., funded by the UK MRC, Alzheimer's Society, and Alzheimer's Research UK. She has undertaken consultancy services for Alnylam Pharmaceuticals Inc., Atalanta Pharmaceuticals (SAB), F. Hoffmann-La Roche Ltd/ Genentech, Guidepoint, Horama, Locanobio, LoQus23 Therapeutics Ltd (SAB), Novartis Pharma, PTC Therapeutics, Sanofi, Spark Therapeutics, Takeda Pharmaceuticals Ltd, Triplet Therapeutics (SAB), University College Irvine, Vertex Pharmaceuticals Incorporated and Wave Life Sciences. All honoraria for these consultancies were paid through the offices of UCL Consultants Ltd., a wholly owned subsidiary of University College London. S.J.T has a patent Application number 2105484.6 on the FAN1-MLH1 interaction and structural analogues licensed to Adrestia Therapeutics. A.D. serves on the advisory boards of Triplet Therapeutics and Minoryx Therapeutics. She holds partly a Patent B 06291873.5 ‘Anaplerotic therapy of Huntington's disease and other polyglutamine diseases’. B.L. has provided consulting services, advisory board functions, clinical trial services, and/or lectures for Acadia Pharmaceuticals, Affiris, Allergan, Alnylam, Amarin, AOP Orphan Pharmaceuticals AG, Bayer Pharma AG, Boehringer-Ingelheim, CHDI Foundation, Deutsche Huntington-Hilfe, Desitin, Genentech, Genzyme, GlaxoSmithKline, F. Hoffmann-La Roche, Ipsen, ISIS Pharma (IONIS), Lilly, Lundbeck, Medesis, Medivation, Medtronic, NeuraMetrix, Neurosearch Inc., Novartis, Pfizer, Prana Biotechnology, Prilenia, PTC Therapeutics, Raptor, Remix Therapeutics, Rhône-Poulenc Rorer, Roche Pharma AG Deutschland, Sage Therapeutics, Sanofi-Aventis, Sangamo/Shire, Siena Biotech, Takeda, Temmler Pharma GmbH, Teva, Triplet TX, Trophos, UniQure, and Wave Life Sciences; he has received research grant support from the CHDI Foundation, the Bundesministerium für Bildung und Forschung (BMBF), the Deutsche Forschungsgemeinschaft (DFG), the European Commission (EU-FP7), EU Joint Programme—Neurodegenerative Disease Research (JNPD), and ERA-Net for Research Programmes on Rare Diseases (E-Rare); his study site has received compensation in the context of the observational REGISTRY-Study of European Huntington's Disease Network (EHDN) and the global observational Enroll-HD; in the context of clinical trials, his institution, the University Hospital of Ulm, has received compensation from Allergan, Ionis, F. Hoffmann-La Roche, Pfizer, and Teva. B.R.L is on the Scientific Advisory Board of sRNAlytics (GateHouse Bio) for which he received stock options, and reports scientific consultancy fees from Teva, Roche/Genentech, Takeda, Triplet, Ionis, Novartis, Spark, Scintetica, LifeEdit, Design, Remix Therapeutics, and PTC Therapeutics. B.R.L.’s laboratory has obtained previous and current research grants from CIHR, HSC, NMIN, CHDI, Teva, ProMIS and uniQure. He is a founding co-Editor-in-Chief,
Figures

Comment in
-
Unravelling the role of huntingtin: from neurodevelopment to neurodegeneration.Brain. 2023 Nov 2;146(11):4408-4410. doi: 10.1093/brain/awad353. Brain. 2023. PMID: 37816304 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

