Rare disease variant curation from literature: assessing gaps with creatine transport deficiency in focus
- PMID: 37587458
- PMCID: PMC10433598
- DOI: 10.1186/s12864-023-09561-5
Rare disease variant curation from literature: assessing gaps with creatine transport deficiency in focus
Abstract
Background: Approximately 4-8% of the world suffers from a rare disease. Rare diseases are often difficult to diagnose, and many do not have approved therapies. Genetic sequencing has the potential to shorten the current diagnostic process, increase mechanistic understanding, and facilitate research on therapeutic approaches but is limited by the difficulty of novel variant pathogenicity interpretation and the communication of known causative variants. It is unknown how many published rare disease variants are currently accessible in the public domain.
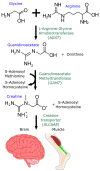
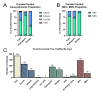
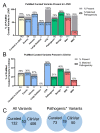
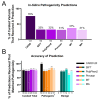
Results: This study investigated the translation of knowledge of variants reported in published manuscripts to publicly accessible variant databases. Variants, symptoms, biochemical assay results, and protein function from literature on the SLC6A8 gene associated with X-linked Creatine Transporter Deficiency (CTD) were curated and reported as a highly annotated dataset of variants with clinical context and functional details. Variants were harmonized, their availability in existing variant databases was analyzed and pathogenicity assignments were compared with impact algorithm predictions. 24% of the pathogenic variants found in PubMed articles were not captured in any database used in this analysis while only 65% of the published variants received an accurate pathogenicity prediction from at least one impact prediction algorithm.
Conclusions: Despite being published in the literature, pathogenicity data on patient variants may remain inaccessible for genetic diagnosis, therapeutic target identification, mechanistic understanding, or hypothesis generation. Clinical and functional details presented in the literature are important to make pathogenicity assessments. Impact predictions remain imperfect but are improving, especially for single nucleotide exonic variants, however such predictions are less accurate or unavailable for intronic and multi-nucleotide variants. Developing text mining workflows that use natural language processing for identifying diseases, genes and variants, along with impact prediction algorithms and integrating with details on clinical phenotypes and functional assessments might be a promising approach to scale literature mining of variants and assigning correct pathogenicity. The curated variants list created by this effort includes context details to improve any such efforts on variant curation for rare diseases.
Keywords: CTD; Gene variant; Literature curation; Rare disease; SLC6A8; Text mining; Variant database.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Group L. The National Economic Burden of Rare Disease Study. EveryLife Foundation for Rare Diseases; 2021.
-
- Ferreira CR, Med Genet A. 2019;179(6):885–92. Epub 2019/03/19. 10.1002/ajmg.a.61124. PubMed PMID: 30883013. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

