Ubiquitination in the regulation of autophagy
- PMID: 37587758
- PMCID: PMC10520486
- DOI: 10.3724/abbs.2023149
Ubiquitination in the regulation of autophagy
Abstract
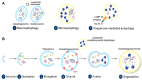
Autophagy, an efficient and effective approach to clear rapidly damaged organelles, macromolecules, and other harmful cellular components, enables the recycling of nutrient materials and supply of nutrients to maintain cellular homeostasis. Ubiquitination plays an important regulatory role in autophagy. This paper summarizes the most recent progress in ubiquitin modification in various stages of autophagy, including initiation, elongation, and termination. Moreover, this paper shows that ubiquitination is an important way through which selective autophagy achieves substrate specificity. Furthermore, we note the distinction between monoubiquitination and polyubiquitination in the regulation of autophagy. Compared with monoubiquitination, polyubiquitination is a more common strategy to regulate the activity of the autophagy molecular machinery. In addition, the role of ubiquitination in the closure and fusion of autophagosomes warrants further study. This article not only clarifies the regulatory mechanism of autophagy but also contributes to a deeper understanding of the importance of ubiquitination modification.
Keywords: autophagy molecule machine; monoubiquitination; polyubiquitination; selective autophagy.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. . 1992;119:301–311. doi: 10.1083/jcb.119.2.301. - DOI - PMC - PubMed
-
- Lamark T, Johansen T. Mechanisms of selective autophagy. Annu Rev Cell Dev Biol. . 2021;37:143–169. doi: 10.1146/annurev-cellbio-120219-035530. - DOI - PubMed
-
- Harper JW, Ordureau A, Heo JM. Building and decoding ubiquitin chains for mitophagy. Nat Rev Mol Cell Biol. . 2018;19:93–108. doi: 10.1038/nrm.2017.129. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

