The making of the oral microbiome in Agta hunter-gatherers
- PMID: 37587941
- PMCID: PMC10426117
- DOI: 10.1017/ehs.2023.9
The making of the oral microbiome in Agta hunter-gatherers
Abstract
Ecological and genetic factors have influenced the composition of the human microbiome during our evolutionary history. We analysed the oral microbiota of the Agta, a hunter-gatherer population where some members have adopted an agricultural diet. We show that age is the strongest factor modulating the microbiome, probably through immunosenescence since we identified an increase in the number of species classified as pathogens with age. We also characterised biological and cultural processes generating sexual dimorphism in the oral microbiome. A small subset of oral bacteria is influenced by the host genome, linking host collagen genes to bacterial biofilm formation. Our data also suggest that shifting from a fish/meat diet to a rice-rich diet transforms their microbiome, mirroring the Neolithic transition. All of these factors have implications in the epidemiology of oral diseases. Thus, the human oral microbiome is multifactorial and shaped by various ecological and social factors that modify the oral environment.
Keywords: Neolithic transition; hunter–gatherers; oral microbiome; pathogen transmission.
© The Author(s) 2023.
Conflict of interest statement
The authors declare none.
Figures

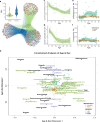
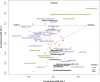

References
-
- Adler, C. J., Dobney, K., Weyrich, L. S., Kaidonis, J., Walker, A. W., Haak, W., … Cooper, A. (2013). Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nature Genetics, 45(4), 450–455. 10.1038/ng.2536 - DOI - PMC - PubMed
-
- Alexa, A. & Rahnenfuhrer, J. (2019). topGO: Enrichment Analysis for Gene Ontology. R package version 2.52.0.
-
- Bellussi, L., Passali, F. M., Ralli, M., De Vincentiis, M., Greco, A. V., & Passali, D. (2019). An overview on upper respiratory tract infections and bacteriotherapy as innovative therapeutic strategy. European Reviews on Medical and Pharmacological Science, 23, 27–38. 10.26355/eurrev_201903_17345 - DOI - PubMed
LinkOut - more resources
Full Text Sources
