Increased KRAS G12C Prevalence, High Tumor Mutational Burden, and Specific Mutational Signatures Are Associated With MUTYH Mutations: A Pan-Cancer Analysis
- PMID: 37589222
- PMCID: PMC10836311
- DOI: 10.1093/oncolo/oyad230
Increased KRAS G12C Prevalence, High Tumor Mutational Burden, and Specific Mutational Signatures Are Associated With MUTYH Mutations: A Pan-Cancer Analysis
Abstract
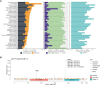
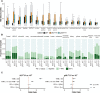
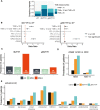
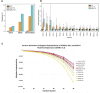
The aim of this study was to determine the pan-cancer landscape of MUTYH alterations and the relationship between MUTYH mutations and potentially actionable biomarkers such as specific genomic alterations, tumor mutational burden, and mutational signatures. We used a large pan-cancer comprehensive genomic dataset from patients profiled (tissue next generation sequencing) during routine clinical care. Overall, 2.8% of 229 120 solid tumors had MUTYH alterations, of which 55% were predicted germline. Thirty tumor types had a 2% or greater MUTYH mutation rate. MUTYH-altered versus -WT cancers had significantly higher tumor mutational burden and more frequent alterations in KRAS G12C, but not in KRAS in general; these observations were statistically significant, especially in colorectal cancers. Across cancers, PD-L1 expression levels (immunohistochemistry) were not associated with MUTYH alteration status. In silico computation demonstrated that MUTYH mutational signatures are associated with higher levels of hydrophobicity (which may reflect higher immunogenicity of neoantigens) relative to several other signature types such as microsatellite instability. Survival of patients with MUTYH-altered versus -WT tumors was similar. In conclusion, comprehensive genomic profiling suggests that several features of MUTYH-altered cancers may be pharmacologically targetable. Drugs such as sotorasib (targeting KRAS G12C) and immune checkpoint inhibitors, targeting the increased mutational load and higher neo-antigen hydrophobicity/immunogenicity merit investigation in MUTYH-mutated malignancies.
Keywords: KRAS G12C; MUTYH mutations; genomic alterations; tumor mutational burden.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
Umut Disel: Foundation Medicine as consultant, speaker fees, and/or advisory board. Smruthy Sivakumar: Employee at Foundation Medicine, a wholly owned subsidiary of Roche and a shareholder in Roche. Zoe Fleischmann: Employee at Foundation Medicine, a wholly owned subsidiary of Roche and a shareholder in Roche. Ethan S. Sokol: Employee at Foundation Medicine, a wholly owned subsidiary of Roche and a shareholder in Roche. Razelle Kurzrock: has received research funding from Biological Dynamics, Boehringer Ingelheim, Debiopharm, Foundation Medicine, Genentech, Grifols, Guardant, Incyte, Konica Minolta, Medimmune, Merck Serono, Omniseq, Pfizer, Sequenom, Takeda, and TopAlliance; as well as consultant and/or speaker fees and/or advisory board for Actuate Therapeutics, AstraZeneca, Bicara Therapeutics, Biological Dynamics, Daiichi Sankyo, Inc., EISAI, EOM Pharmaceuticals, Iylon, Merck, NeoGenomics, Neomed, Pfizer, Prosperdtx, Roche, TD2/Volastra, Turning Point Therapeutics, X-Biotech; has an equity interest in CureMatch Inc., CureMetrix, and IDbyDNA; serves on the Board of CureMatch and CureMetrix, and is a co-founder of CureMatch. The other authors indicated no financial relationships.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

