Infection and tissue distribution of highly pathogenic avian influenza A type H5N1 (clade 2.3.4.4b) in red fox kits (Vulpes vulpes)
- PMID: 37589241
- PMCID: PMC10512766
- DOI: 10.1080/22221751.2023.2249554
Infection and tissue distribution of highly pathogenic avian influenza A type H5N1 (clade 2.3.4.4b) in red fox kits (Vulpes vulpes)
Abstract
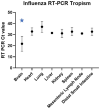
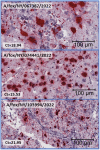
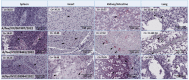
Avian influenza H5N1 is a highly pathogenic virus that primarily affects birds. However, it can also infect other animal species, including mammals. We report the infection of nine juvenile red foxes (Vulpes vulpes) with Highly Pathogenic Avian Influenza A type H5N1 (Clade 2.3.4.4b) in the spring of 2022 in the central, western, and northern regions of New York, USA. The foxes displayed neurologic signs, and examination of brain and lung tissue revealed lesions, with brain lesions ranging from moderate to severe meningoencephalitis. Analysis of tissue tropism using RT-PCR methods showed a comparatively lower Ct value in the brain, which was confirmed by in situ hybridization targeting Influenza A RNA. The viral RNA labelling was highly clustered and overlapped the brain lesions, observed in neurons, and grey matter. Whole viral genome sequences obtained from the affected foxes were subjected to phylogenetic and mutation analysis to determine influenza A clade, host specificity, and potential occurrence of viral reassortment. Infections in red foxes likely occurred due to preying on infected wild birds and are unlikely due to transmission between foxes or other mammals.
Keywords: 2.3.4.4b; H5N1; Influenza; Vulpes vulpes; highly pathogenic avian influenza A; red fox; tissue tropism; whole genome sequencing.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




References
-
- Böttcher-Friebertshäuser E, Garten W, Matrosovich M, et al. The hemagglutinin: a determinant of pathogenicity. In: Compans RW, Oldstone MBA, editor. Influenza pathogenesis and control – volume I [Internet]. Cham: Springer International Publishing; 2014. [cited 2023 Jun 20]. p. 3–34. DOI: 10.1007/82_2014_384 - DOI - PubMed
-
- Sagong M, Lee Y, Song S, et al. Emergence of clade 2.3.4.4b novel reassortant H5N1 high pathogenicity avian influenza virus in South Korea during late 2021. Transbounding Emerg Dis. 2022:tbed.14551. - PubMed
-
- Bevins SN, Shriner SA, Cumbee JC, et al. Intercontinental movement of H5 2.3.4.4 highly pathogenic avian influenza A (H5N1) to the United States, 2021 [Internet]. Microbiology. 2022 [cited 2022 Jul 13]. Available from: http://biorxiv.org/lookup/doi/10.11012022.02.11.479922 - DOI - PMC - PubMed
-
- Richards B. Distribution of highly pathogenic avian influenza in North America, 2021/2022 [Internet]. [cited 2022 Jul 18]. Available from: https://www.usgs.gov/centers/nwhc/science/distribution-highly-pathogenic...
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
