ARA: a flexible pipeline for automated exploration of NCBI SRA datasets
- PMID: 37589306
- PMCID: PMC10433097
- DOI: 10.1093/gigascience/giad067
ARA: a flexible pipeline for automated exploration of NCBI SRA datasets
Abstract
Background: One of the most effective and useful methods to explore the content of biological databases is searching with nucleotide or protein sequences as a query. However, especially in the case of nucleic acids, due to the large volume of data generated by the next-generation sequencing (NGS) technologies, this approach is often not available. The hierarchical organization of the NGS records is primarily designed for browsing or text-based searches of the information provided in metadata-related keywords, limiting the efficiency of database exploration.
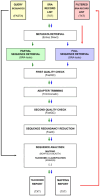
Findings: We developed an automated pipeline that incorporates the well-established NGS data-processing tools and procedures to allow easy and effective sampling of the NCBI SRA database records. Given a file with query nucleotide sequences, our tool estimates the matching content of SRA accessions by probing only a user-defined fraction of a record's sequences. Based on the selected parameters, it allows performing a full mapping experiment with records that meet the required criteria. The pipeline is designed to be easy to operate-it offers a fully automatic setup procedure and is fixed on tested supporting tools. The modular design and implemented usage modes allow a user to scale up the analyses into complex computational infrastructure.
Conclusions: We present an easy-to-operate and automated tool that expands the way a user can access and explore the information contained within the records deposited in the NCBI SRA database.
Keywords: NGS data; SRA database; database searching; sequence analysis.
© The Author(s) 2023. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

