AutophagyNet: high-resolution data source for the analysis of autophagy and its regulation
- PMID: 37589496
- PMCID: PMC10761021
- DOI: 10.1080/15548627.2023.2247737
AutophagyNet: high-resolution data source for the analysis of autophagy and its regulation
Abstract
Macroautophagy/autophagy is a highly-conserved catabolic procss eliminating dysfunctional cellular components and invading pathogens. Autophagy malfunction contributes to disorders such as cancer, neurodegenerative and inflammatory diseases. Understanding autophagy regulation in health and disease has been the focus of the last decades. We previously provided an integrated database for autophagy research, the Autophagy Regulatory Network (ARN). For the last eight years, this resource has been used by thousands of users. Here, we present a new and upgraded resource, AutophagyNet. It builds on the previous database but contains major improvements to address user feedback and novel needs due to the advancement in omics data availability. AutophagyNet contains updated interaction curation and integration of over 280,000 experimentally verified interactions between core autophagy proteins and their protein, transcriptional and post-transcriptional regulators as well as their potential upstream pathway connections. AutophagyNet provides annotations for each core protein about their role: 1) in different types of autophagy (mitophagy, xenophagy, etc.); 2) in distinct stages of autophagy (initiation, expansion, termination, etc.); 3) with subcellular and tissue-specific localization. These annotations can be used to filter the dataset, providing customizable download options tailored to the user's needs. The resource is available in various file formats (e.g. CSV, BioPAX and PSI-MI), and data can be analyzed and visualized directly in Cytoscape. The multi-layered regulation of autophagy can be analyzed by combining AutophagyNet with tissue- or cell type-specific (multi-)omics datasets (e.g. transcriptomic or proteomic data). The resource is publicly accessible at http://autophagynet.org.Abbreviations: ARN: Autophagy Regulatory Network; ATG: autophagy related; BCR: B cell receptor pathway; BECN1: beclin 1; GABARAP: GABA type A receptor-associated protein; IIP: innate immune pathway; LIR: LC3-interacting region; lncRNA: long non-coding RNA; MAP1LC3B: microtubule associated protein 1 light chain 3 beta; miRNA: microRNA; NHR: nuclear hormone receptor; PTM: post-translational modification; RTK: receptor tyrosine kinase; TCR: T cell receptor; TLR: toll like receptor.
Keywords: Autophagy regulation; big data; multi-omics; network resource; signaling.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

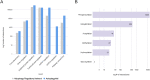




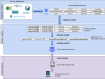
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
