Cooperative regulation of C1-domain membrane recruitment polarizes atypical protein kinase C
- PMID: 37589718
- PMCID: PMC10435729
- DOI: 10.1083/jcb.202112143
Cooperative regulation of C1-domain membrane recruitment polarizes atypical protein kinase C
Abstract
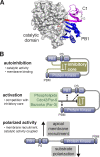
Recruitment of the Par complex protein atypical protein kinase C (aPKC) to a specific membrane domain is a key step in the polarization of animal cells. While numerous proteins and phospholipids interact with aPKC, how these interactions cooperate to control its membrane recruitment has been unknown. Here, we identify aPKC's C1 domain as a phospholipid interaction module that targets aPKC to the membrane of Drosophila neural stem cells (NSCs). The isolated C1 binds the NSC membrane in an unpolarized manner during interphase and mitosis and is uniquely sufficient among aPKC domains for targeting. Other domains, including the catalytic module and those that bind the upstream regulators Par-6 and Bazooka, restrict C1's membrane targeting activity-spatially and temporally-to the apical NSC membrane during mitosis. Our results suggest that aPKC polarity results from cooperative activation of autoinhibited C1-mediated membrane binding activity.
© 2023 Jones et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

