Feasibility and clinical utility of local rapid Nanopore influenza A virus whole genome sequencing for integrated outbreak management, genotypic resistance detection and timely surveillance
- PMID: 37590039
- PMCID: PMC10483427
- DOI: 10.1099/mgen.0.001083
Feasibility and clinical utility of local rapid Nanopore influenza A virus whole genome sequencing for integrated outbreak management, genotypic resistance detection and timely surveillance
Abstract
Rapid respiratory viral whole genome sequencing (WGS) in a clinical setting can inform real-time outbreak and patient treatment decisions, but the feasibility and clinical utility of influenza A virus (IAV) WGS using Nanopore technology has not been demonstrated. A 24 h turnaround Nanopore IAV WGS protocol was performed on 128 reverse transcriptase PCR IAV-positive nasopharyngeal samples taken over seven weeks of the 2022-2023 winter influenza season, including 25 from patients with nosocomial IAV infections and 102 from patients attending the Emergency Department. WGS results were reviewed collectively alongside clinical details for interpretation and reported to clinical teams. All eight segments of the IAV genome were recovered for 97/128 samples (75.8 %) and the haemagglutinin gene for 117/128 samples (91.4 %). Infection prevention and control identified nosocomial IAV infections in 19 patients across five wards. IAV WGS revealed two separate clusters on one ward and excluded transmission across different wards with contemporaneous outbreaks. IAV WGS also identified neuraminidase inhibitor resistance in a persistently infected patient and excluded avian influenza in a sample taken from an immunosuppressed patient with a history of travel to Singapore which had failed PCR subtyping. Accurate IAV genomes can be generated in 24 h using a Nanopore protocol accessible to any laboratory with SARS-CoV-2 Nanopore sequencing capacity. In addition to replicating reference laboratory surveillance results, IAV WGS can identify antiviral resistance and exclude avian influenza. IAV WGS also informs management of nosocomial outbreaks, though molecular and clinical epidemiology were concordant in this study, limiting the impact on decision-making.
Keywords: Nanopore; antiviral drug resistance; avian influenza; influenza A; nosocomial outbreaks; whole genome sequencing.
Conflict of interest statement
AA-M and JKS are full-time employees of Synnovis. JDE holds part-time employment contract with Oxford Nanopore Technologies as VP Medical Affairs. Guy’s and St Thomas’ NHS Foundation Trust signed a commercial collaboration agreement with Oxford Nanopore Technology in September 2022. TGSW, LBS, CA, TC, NA-Y, GH, SM, AL, MZ, SD and GN have no conflicts of interest to declare.
Figures
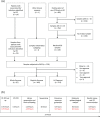


References
-
- UK Health Security Agency Weekly national Influenza and COVID-19 surveillance report. Week 51 report. [ January 29; 2023 ];2022 https://assets.publishing.service.gov.uk/government/uploads/system/uploa... accessed.
-
- Meredith LW, Hamilton WL, Warne B, Houldcroft CJ, Hosmillo M, et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis. 2020;20:1263–1272. doi: 10.1016/S1473-3099(20)30562-4. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

