The C9ORF72 repeat expansion alters neurodevelopment
- PMID: 37590144
- PMCID: PMC10757587
- DOI: 10.1016/j.celrep.2023.112983
The C9ORF72 repeat expansion alters neurodevelopment
Abstract
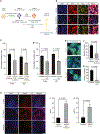
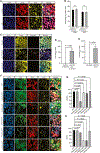
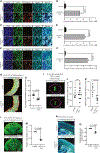
Genetic mutations that cause adult-onset neurodegenerative diseases are often expressed during embryonic stages, but it is unclear whether they alter neurodevelopment and how this might influence disease onset. Here, we show that the most common cause of frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS), a repeat expansion in C9ORF72, restricts neural stem cell proliferation and reduces cortical and thalamic size in utero. Surprisingly, a repeat expansion-derived dipeptide repeat protein (DPR) not known to reduce neuronal viability plays a key role in impairing neurodevelopment. Pharmacologically mimicking the effects of the repeat expansion on neurodevelopment increases susceptibility of C9ORF72 mice to motor defects. Thus, the C9ORF72 repeat expansion stunts development of the brain regions prominently affected in C9ORF72 FTD/ALS patients.
Keywords: C9ORF72; CP: Neuroscience; amyotrophic lateral sclerosis; frontotemporal dementia; induced pluripotent stem cells; neural stem cells; neurodevelopment.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.K.I. is a co-founder of AcuraStem, Inc. and Modulo Bio. J.K.I. is on the scientific advisory board of AcuraStem, Inc.; Spinogenix; and Vesalius Therapeutics. J.K.I. is an employee of BioMarin Pharmaceutical. J.K.I. is a co-founder of Modulo Bio and serves on the scientific advisory board of Spinogenix. Named companies were not involved in this project.
Figures


References
-
- Deneubourg C, Ramm M, Smith LJ, Baron O, Singh K, Byrne SC,Duchen MR, Gautel M, Eskelinen E-L, Fanto M, and Jungbluth H. (2022). The spectrum of neurodevelopmental, neuromuscular and neurodegenerative disorders due to defective autophagy. Autophagy 18, 496–517. 10.1080/15548627.2021.1943177. - DOI - PMC - PubMed
-
- Arteaga-Bracho EE, Gulinello M, Winchester ML, Pichamoorthy N, Petronglo JR, Zambrano AD, Inocencio J, De Jesus CD, Louie JO, Gokhan S, et al. (2016). Postnatal and adult consequences of loss of huntingtin during development: Implications for Huntington’s disease. Neurobiol. Dis. 96, 144–155. 10.1016/j.nbd.2016.09.006. - DOI - PMC - PubMed
-
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. (2011). Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256. 10.1016/j.neuron.2011.09.011. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

