Multidimensional scaling methods can reconstruct genomic DNA loops using Hi-C data properties
- PMID: 37590265
- PMCID: PMC10434948
- DOI: 10.1371/journal.pone.0289651
Multidimensional scaling methods can reconstruct genomic DNA loops using Hi-C data properties
Abstract
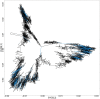
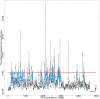
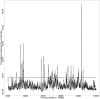
This paper proposes multidimensional scaling (MDS) applied to high-throughput chromosome conformation capture (Hi-C) data on genomic interactions to visualize DNA loops. Currently, the mechanisms underlying the regulation of gene expression are poorly understood, and where and when DNA loops are formed remains undetermined. Previous studies have focused on reproducing the entire three-dimensional structure of chromatin; however, identifying DNA loops using these data is time-consuming and difficult. MDS is an unsupervised method for reconstructing the original coordinates from a distance matrix. Here, MDS was applied to high-throughput chromosome conformation capture (Hi-C) data on genomic interactions to visualize DNA loops. Hi-C data were converted to distances by taking the inverse to reproduce loops via MDS, and the missing values were set to zero. Using the converted data, MDS was applied to the log-transformed genomic coordinate distances and this process successfully reproduced the DNA loops in the given structure. Consequently, the reconstructed DNA loops revealed significantly more DNA-transcription factor interactions involved in DNA loop formation than those obtained from previously applied methods. Furthermore, the reconstructed DNA loops were significantly consistent with chromatin immunoprecipitation followed by sequencing (ChIP-seq) peak positions. In conclusion, the proposed method is an improvement over previous methods for identifying DNA loops.
Copyright: © 2023 Ryo Ishibashi. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

