An ATR-PrimPol pathway confers tolerance to oncogenic KRAS-induced and heterochromatin-associated replication stress
- PMID: 37591859
- PMCID: PMC10435487
- DOI: 10.1038/s41467-023-40578-2
An ATR-PrimPol pathway confers tolerance to oncogenic KRAS-induced and heterochromatin-associated replication stress
Abstract
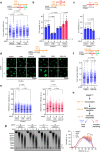
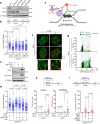
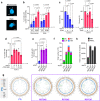
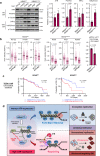
Activation of the KRAS oncogene is a source of replication stress, but how this stress is generated and how it is tolerated by cancer cells remain poorly understood. Here we show that induction of KRASG12V expression in untransformed cells triggers H3K27me3 and HP1-associated chromatin compaction in an RNA transcription dependent manner, resulting in replication fork slowing and cell death. Furthermore, elevated ATR expression is necessary and sufficient for tolerance of KRASG12V-induced replication stress to expand replication stress-tolerant cells (RSTCs). PrimPol is phosphorylated at Ser255, a potential Chk1 substrate site, under KRASG12V-induced replication stress and promotes repriming to maintain fork progression and cell survival in an ATR/Chk1-dependent manner. However, ssDNA gaps are generated at heterochromatin by PrimPol-dependent repriming, leading to genomic instability. These results reveal a role of ATR-PrimPol in enabling precancerous cells to survive KRAS-induced replication stress and expand clonally with accumulation of genomic instability.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability–an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010;11:220–228. - PubMed
-
- Macheret M, Halazonetis TD. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 2015;10:425–448. - PubMed
-
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

