C16orf72/HAPSTR1/TAPR1 functions with BRCA1/Senataxin to modulate replication-associated R-loops and confer resistance to PARP disruption
- PMID: 37591890
- PMCID: PMC10435583
- DOI: 10.1038/s41467-023-40779-9
C16orf72/HAPSTR1/TAPR1 functions with BRCA1/Senataxin to modulate replication-associated R-loops and confer resistance to PARP disruption
Erratum in
-
Author Correction: C16orf72/HAPSTR1/TAPR1 functions with BRCA1/Senataxin to modulate replication-associated R-loops and confer resistance to PARP disruption.Nat Commun. 2023 Nov 27;14(1):7784. doi: 10.1038/s41467-023-43353-5. Nat Commun. 2023. PMID: 38012134 Free PMC article. No abstract available.
Abstract
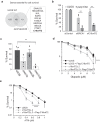
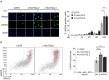
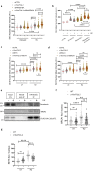
While the toxicity of PARP inhibitors to cells with defects in homologous recombination (HR) is well established, other synthetic lethal interactions with PARP1/PARP2 disruption are poorly defined. To inform on these mechanisms we conducted a genome-wide screen for genes that are synthetic lethal with PARP1/2 gene disruption and identified C16orf72/HAPSTR1/TAPR1 as a novel modulator of replication-associated R-loops. C16orf72 is critical to facilitate replication fork restart, suppress DNA damage and maintain genome stability in response to replication stress. Importantly, C16orf72 and PARP1/2 function in parallel pathways to suppress DNA:RNA hybrids that accumulate at stalled replication forks. Mechanistically, this is achieved through an interaction of C16orf72 with BRCA1 and the RNA/DNA helicase Senataxin to facilitate their recruitment to RNA:DNA hybrids and confer resistance to PARP inhibitors. Together, this identifies a C16orf72/Senataxin/BRCA1-dependent pathway to suppress replication-associated R-loop accumulation, maintain genome stability and confer resistance to PARP inhibitors.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

