KDM6A epigenetically regulates subtype plasticity in small cell lung cancer
- PMID: 37591951
- PMCID: PMC10546329
- DOI: 10.1038/s41556-023-01210-z
KDM6A epigenetically regulates subtype plasticity in small cell lung cancer
Abstract
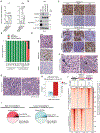
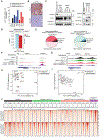
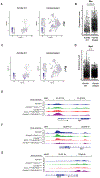
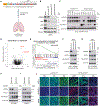
Small cell lung cancer (SCLC) exists broadly in four molecular subtypes: ASCL1, NEUROD1, POU2F3 and Inflammatory. Initially, SCLC subtypes were thought to be mutually exclusive, but recent evidence shows intra-tumoural subtype heterogeneity and plasticity between subtypes. Here, using a CRISPR-based autochthonous SCLC genetically engineered mouse model to study the consequences of KDM6A/UTX inactivation, we show that KDM6A inactivation induced plasticity from ASCL1 to NEUROD1 resulting in SCLC tumours that express both ASCL1 and NEUROD1. Mechanistically, KDM6A normally maintains an active chromatin state that favours the ASCL1 subtype with its loss decreasing H3K4me1 and increasing H3K27me3 at enhancers of neuroendocrine genes leading to a cell state that is primed for ASCL1-to-NEUROD1 subtype switching. This work identifies KDM6A as an epigenetic regulator that controls ASCL1 to NEUROD1 subtype plasticity and provides an autochthonous SCLC genetically engineered mouse model to model ASCL1 and NEUROD1 subtype heterogeneity and plasticity, which is found in 35-40% of human SCLCs.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Figures












Comment in
-
Epigenetic Regulators Open the Door to SCLC Plasticity.Cancer Res. 2023 Nov 1;83(21):3495-3497. doi: 10.1158/0008-5472.CAN-23-2922. Cancer Res. 2023. PMID: 37756567
References
References for Methods
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

